Method for detecting bioactivity of humanized anti-CD52 antibody
A CD52 and antibody biological technology, applied in the biological field, can solve the problems of poor detection method sensitivity, poor detection sensitivity, and difficulty in obtaining a large batch of uniform and sufficient quantities.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1: Construction of CHO-C cells
[0064] 1. Acquisition of human CD52 cDNA and construction of expression vector:
[0065] We designed primer sequences:
[0066] Sense primer: 5′-C CACCATGAAGCGCTTCCTCTTCCTC-3' (SEQ ID NO. 1)
[0067] Antisense primer: 5′-C TCAACTGAAGCAGAAGAGGTG-3' (SEQ ID NO.2)
[0068] This pair of primers has added restriction sites on both sides of the complementary region for loading into the expression vector
[0069] The gene sequence of human CD52 cDNA was obtained from the RNA extract of human peripheral blood by conventional RT-PCR method: 10 ml of normal human peripheral blood was extracted, and mononuclear cells were separated by Ficoll (Beijing Dingguo Biotechnology Co., Ltd.) density gradient centrifugation. Trizol kit (GIBCO company) was used to extract total RNA, and RT-PCR kit (INVITROGEN company) was used for reverse transcription PCR to obtain a specific fragment of about 200 bp. The fragment was recovered with a gel rec...
Embodiment 2
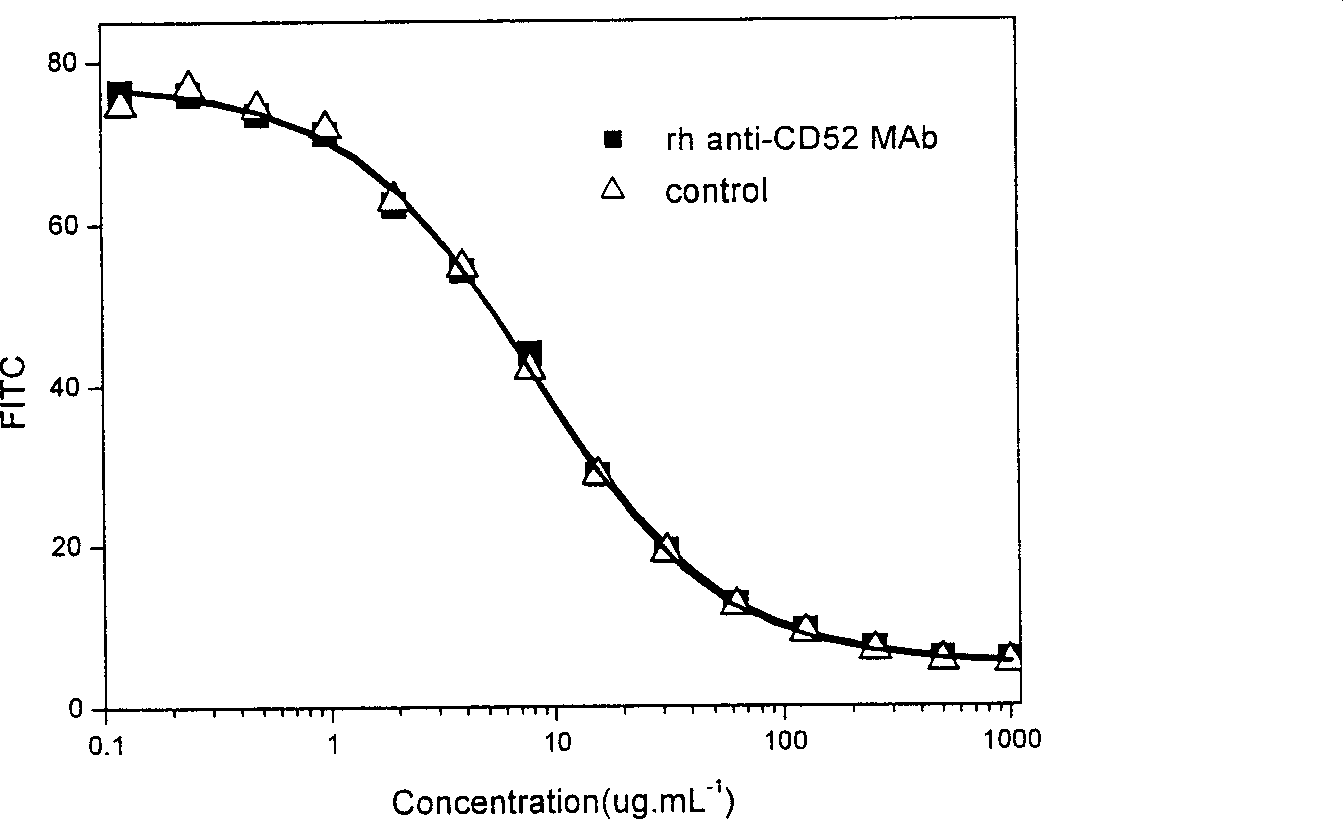
[0076] Embodiment 2: biological activity assay experiment
[0077] 1. Take an aliquoted reference product (Campath-1, product of Genzyme Company), dilute it to 2000 μg / ml with PBSS, and add an equal volume of 20 μg / ml FITC-anti-CD52MAb to each dilution factor of no more than 10 times (PBSS dilution).
[0078] 2. According to the protein quantification of the sample to be tested (that is, rhCD52mAb prepared according to the content disclosed in the Chinese invention patent application number 02155174.X), dilute to 2000 μg / ml with PBSS, and the dilution factor does not exceed 10 times each time, and add an equal volume of 20 μg / ml of FITC-anti-CD52MAb (diluted in PBSS).
[0079] 3. Use 10 μg / ml FITC-anti-CD52MAb (diluted in PBSS) in a centrifuge tube to serially dilute the dilution of the working reference product or the sample by 2 times.
[0080] 4. Take the CHO-C cells in the logarithmic growth phase, count them, and take about 10 for each assay (1 sample and working refer...
experiment example 1
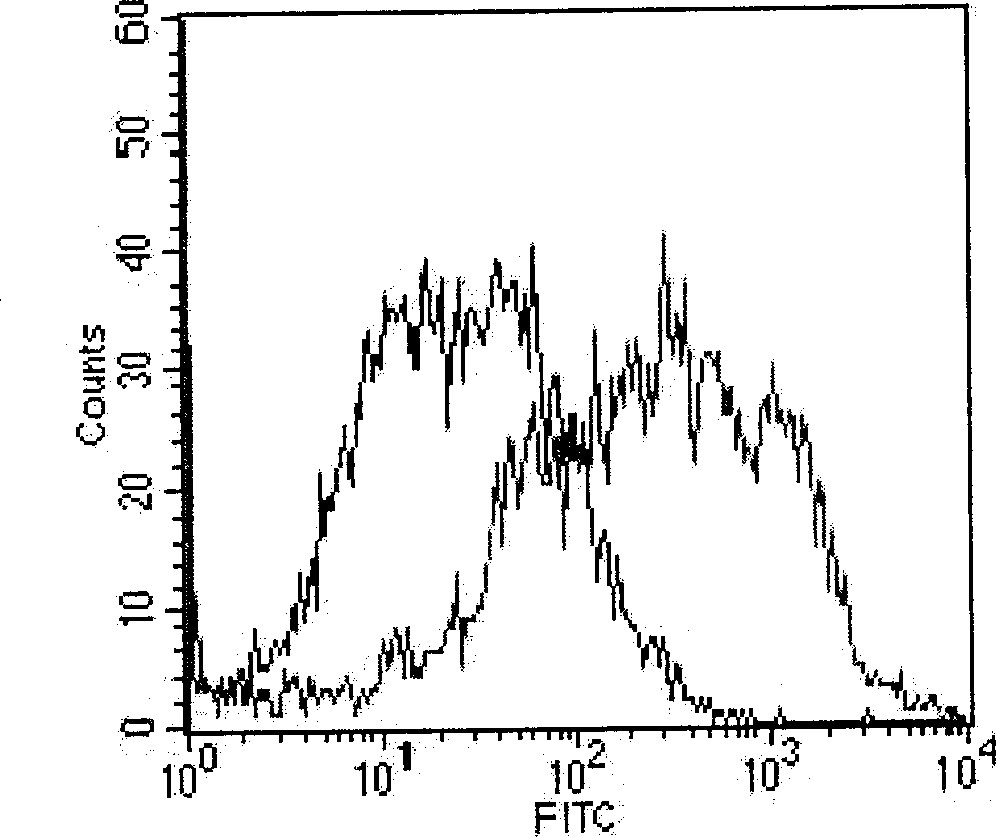
[0091] Experimental Example 1 Identification experiment of CHO-C cells: Refer to "Modern Immunology Experimental Technology" for fluorescent staining
[0092] CHO-K1 cells (ATCC CCL-61) are compared with CHO-cells of the present invention. After labeling, the peak of the staining fluorescence intensity of CHO-C cells can be obviously shifted to the right, that is, FITC-rhCD52mAb can stain CHO-C, that is It shows that CD52 is expressed on the surface of CHO-C cells, but not expressed on the surface of untransfected CHO-K1 cells. The results are detailed in the attached image 3
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com