Method for detecting mercury ion residue of fluorescent signal conversion mechanism based on nucleic acid aptamer structure
A nucleic acid aptamer, fluorescent signal technology, applied in fluorescence/phosphorescence, material excitation analysis, etc., to achieve the effect of small background value, less detection system and sample demand, and strong resistance to matrix interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Construction of mercury ion nucleic acid aptamer N1
[0033] First, according to the base sequence of the nickel ion RNA aptamer reported in the literature, its DNA sequence was synthesized according to the method recorded in the literature Hofmann, HP, Limmer S, Hornung V, Sprinzl M. RNA, 1997, 3: 1289-1300, and its base The sequence is SEQ ID N O .7:
[0034] 5'-AUAGUCAGGGAACAUGACAAACACAGGGACUUGCGAAAAUCAGUGUUUUG-3'.
[0035] The base sequence is defined as DNA1-B.
[0036] When constructing the mercury ion nucleic acid aptamer N1, retain the 23rd to 44th bases of DNA1-B, and replace the 14th base A (equivalent to the 36th base of DNA1-B) with GC ( Add a base here), and finally add the base GG at the 5' end, and at the same time add the base CCG at the 3' end, and replace all the bases U in the reserved segment with the base T, that is, mercury Ionic nucleic acid aptamer N1, its base sequence is SEQ ID N O .2:
[0037] 5'-GGACAGGGACTTGCGGCAAATCAGTCCG-3'.
[0038...
Embodiment 2
[0040] Use the nucleic acid aptamer N1 to establish a fluorescent detection system:
[0041] Synthesize the complementary sequence Q2 with nucleic acid aptamer N1, its base sequence is SEQ ID N O .1:
[0042] 5'-GTCCCTGTCC-3';
[0043] Prepare bufferA buffer: 50mM NaCl+7mM MgCl 2 +50mM Tris / HCl, buffer pH=7.5;
[0044] Fluorescent group FAM, synthesized by Shanghai Jierui Biotechnology Co., Ltd., and it is labeled at the 5' end of the nucleic acid aptamer N1;
[0045] The fluorescence quenching group DABCYL was synthesized by Shanghai Jierui Biotechnology Co., Ltd., and it was labeled at the 3' end of the complementary sequence Q2.
[0046] The nucleic acid aptamer N1 labeled with the fluorescent group FAM and the complementary sequence Q2 labeled with the fluorescent quenching group DABCYL were respectively adjusted at a concentration ratio of 1:0~4 (the final concentration of the nucleic acid aptamer N1 was 50nmol / L, and the complementary sequence Q2 The final concentra...
Embodiment 3
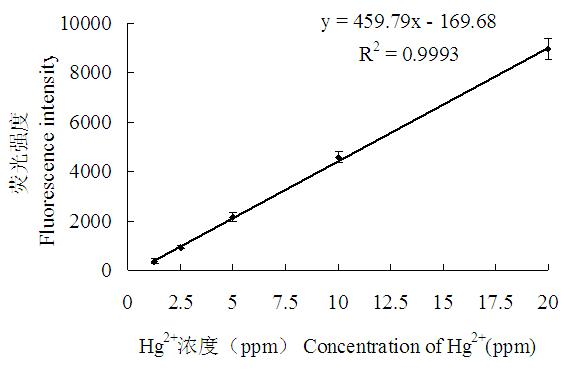
[0051] The detection standard curve of nucleic acid aptamer N1 was established by different concentrations of mercury ions.
[0052] The construction method of the fluorescence detection system is the same as that in Example 2.
[0053] Mix N1 and Q2 in the buffer A buffer system at a ratio of 1:3 (the final concentration of N1 is 150nmol / L, and the final concentration of Q2 is 450nmol / L). Denature at 94°C for 5 minutes, incubate at 25°C for 30 minutes, take out 50 μl from the system, and measure its fluorescence intensity; then add 50 μl of mercury ion standard solution in sequence, and carry out 2-fold gradient dilution of mercury ion concentration from 160ppm to 0.156ppm, And an equal volume of bufferA buffer was used as a control. After 5 minutes, the fluorescence intensity of each well was measured under the excitation light wavelength of 495 nm and the emission light wavelength of 535 nm. Standard curve see figure 2 ,Depend on figure 2 It can be seen that the corre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
| correlation coefficient | aaaaa | aaaaa |
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com