Cooperative reporter systems, components, and methods for analyte detection
An analyte and indicator technology, applied in the direction of analytical materials, biological tests, measuring devices, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1: Peptostreptococcus protein L (PpL) for binding to indicator components at specific sites Modification
[0060] The PpL B1 domain of Peptostreptococcus major has two binding sites for the Ig kappa light chain, which correspond to the two ends of the beta sheet [Svensson et al. 2004, the entire contents of which are hereby incorporated by reference]( Figure 14 ). To form the desired 1:1 ratio of PpL / Ig-κ complexes (as shown in Figure 2), a combination of site-directed mutagenesis and biotin / streptavidin (SA)-mediated protein assembly, was used To block the binding site 1 of Ig-κ. A20C variation can be achieved by site-directed mutagenesis as described, for example, in U.S. Pat. Primer: 5'-TTCCTTTGAATTCGCAAGTTTGTGTGGATCCATTTGC-3') was introduced into the PpLB1 domain containing the Y47W variation (SEQ. ID NO: 1 and 6, provided by Drs. Ulf Sjobring and David Baker). The primer introduces a single cysteine in the center of position 1 [Svensson et al. 200...
Embodiment 2
[0061] Example 2: Modification of Peptostreptococcus protein L (PpL) for covalent attachment to an affinity group decorated
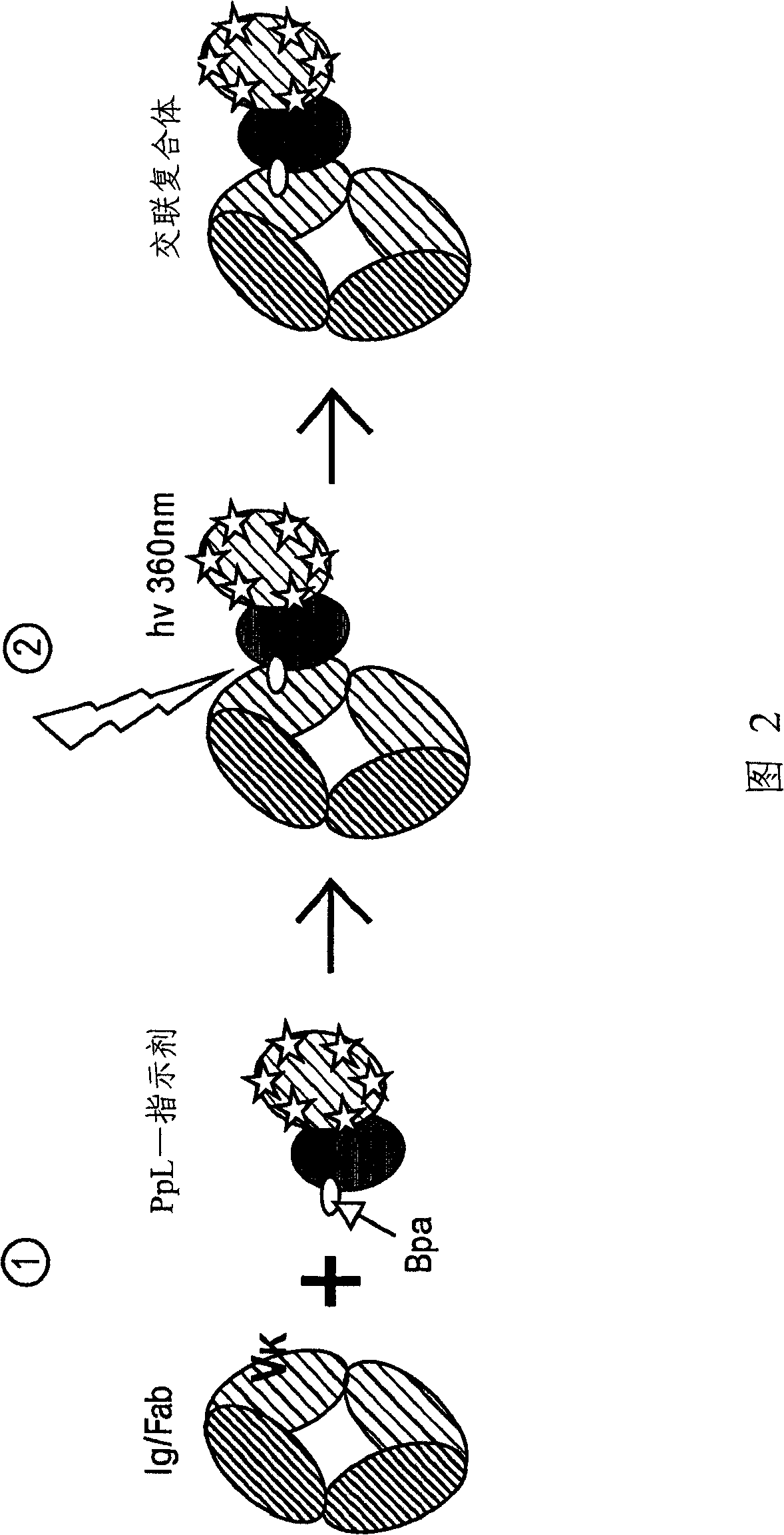
[0062] In order to enable the covalent binding of the PpL B1 domain to the light chain variable region of Igκ, a non-natural photocrosslinking amino acid residue (p-benzoyl-L-phenyl alanine or Bpa) was introduced into a His-tagged PpL B1 domain mutant (Example 1), using an engineered orthogonal aminoacyl-tRNA synthetase / tRNA pair corresponding to the amber codon TAG to convert Bpa in vivo Synthesized into Escherichia coli proteins [Chin et al. into this article]. In the PpL B1 domain structure close to its two binding sites, T39( Figure 14 A is marked in pink) and K41 (in Figure 14 Marked in cyan in B) are where the binding sites 1 and 2 were mutated to Bpa, respectively. For the introduction of Bpa, two plasmids were used: (1) a p15A-based plasmid (pDULE), expressing an orthogonal tRNA and synthetase pair (obtained from Dr. Peter Schultz), an...
Embodiment 3
[0063] Example 3: Attachment of Indicator Components to Modified PpL B1 Molecules
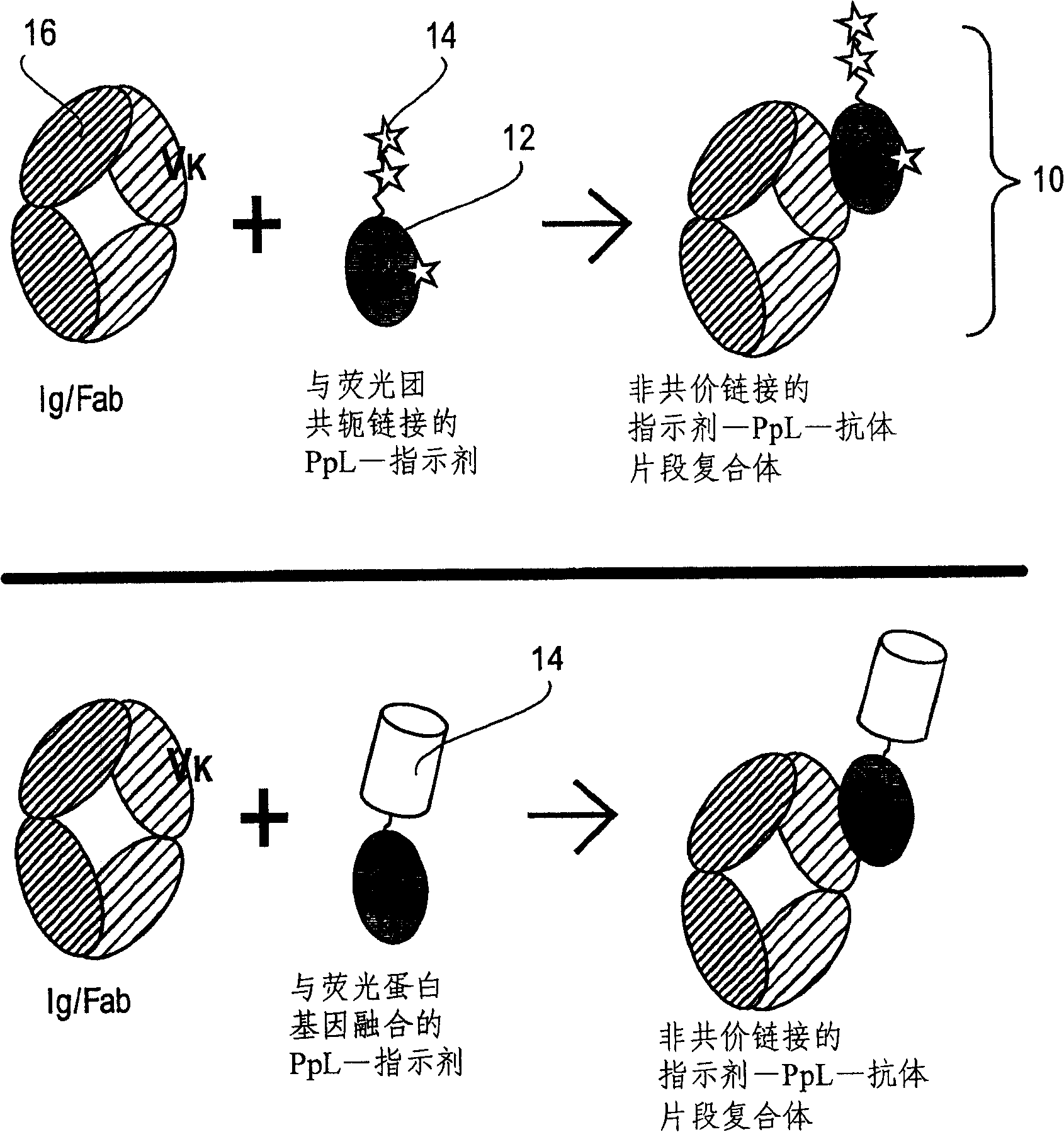
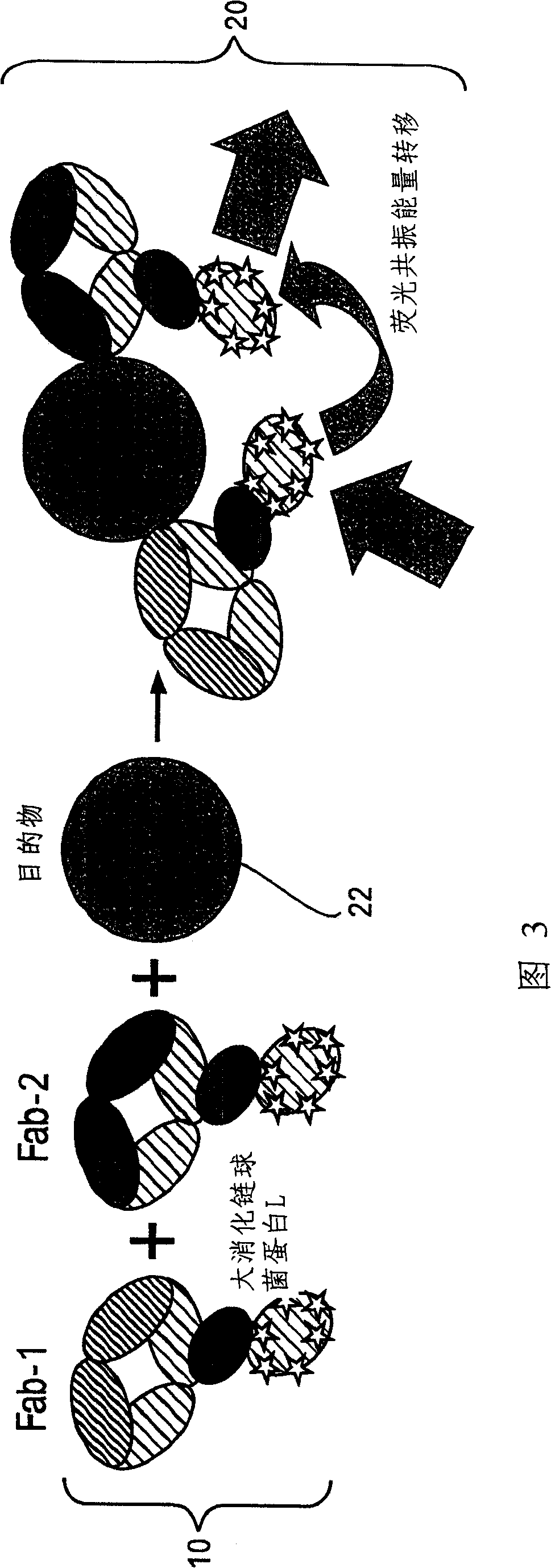
[0064] PpL B1 mutants (Example 1, Example 2) with a biotinylated histidine tag were immobilized in a Ni-NTA column, and fluorophore-labeled streptavidin (SA) was pumped through the column to form a 1:1 complex with the PpL mutant. The protein complexes formed on the column were then washed with imidazole, dialyzed to remove leachable nickel ions and excess imidazole, and subjected to gel filtration chromatography for further purification and analysis to examine their molecular size distribution. In this approach, isolated tubes of PpL B1 domain molecules can be individually labeled to create a variety of adapters for use in assays of the invention.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com