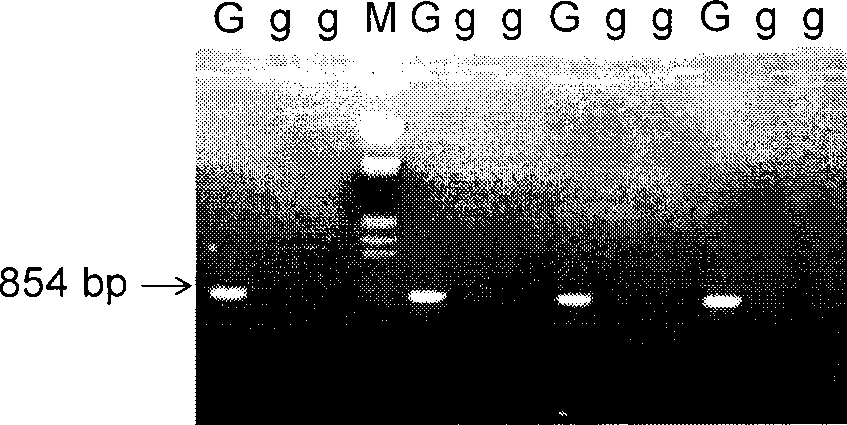Molecular marking method of corn hybrid incompatibly gene
A molecular marker and labeling technology, which is applied in the fields of biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of accelerating the process of variety selection and breeding.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] 1. Construction of Ga gene segregation population
[0042] Taking the Ga inbred line as the parent, crossing with the sweet corn inbred line or the waxy corn inbred line, and constructing Ga gene segregation population F through selfing and backcrossing 2 Generations and BC 1 generation.
[0043] 2. Genomic DNA extraction
[0044] Take 3-4g of young corn leaves and add liquid nitrogen to grind them into powder, put them into a 10mL centrifuge tube, add 5mL of 2×CTAB solution preheated at 65°C, bathe in 65°C for 60min; centrifuge at 5000rpm for 10min, transfer the supernatant to another centrifuge tube; add an equal volume of chloroform / isoamyl alcohol (24:1) and mix lightly; centrifuge at 5000rpm for 10min, transfer the supernatant to another centrifuge tube; add 1 / 100 volume of RNase A solution, and incubate at 37°C for 25min; 0.8 times the volume of isopropanol to precipitate DNA; 2mL (76% ethanol + 0.2mol / L NaAc) to wash the precipitate twice, dry it and dissolve ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com