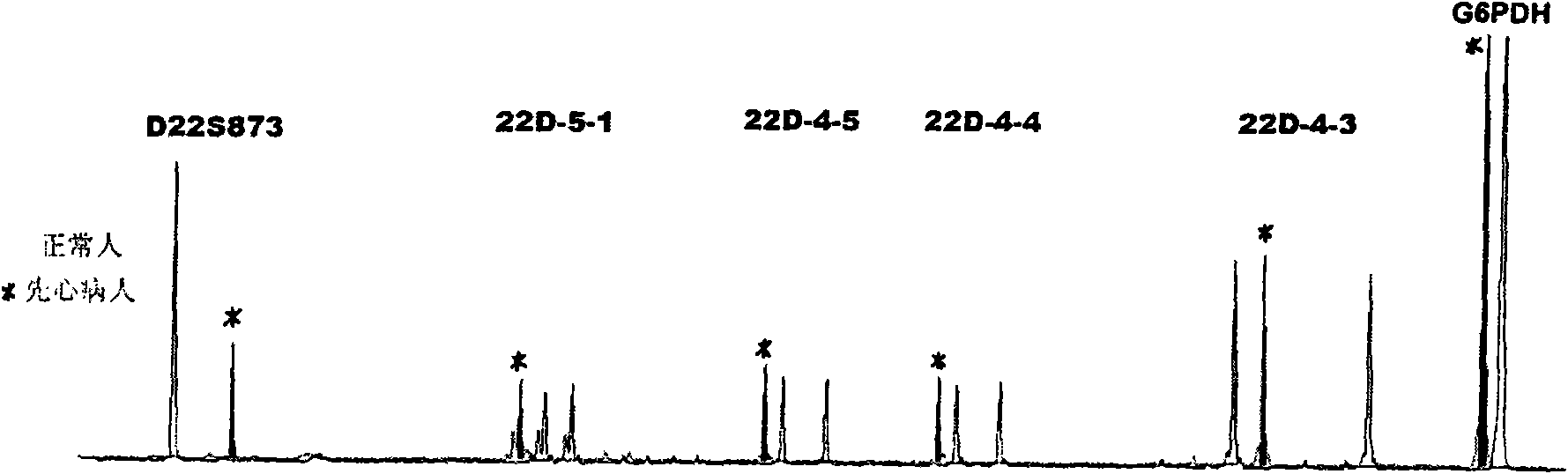
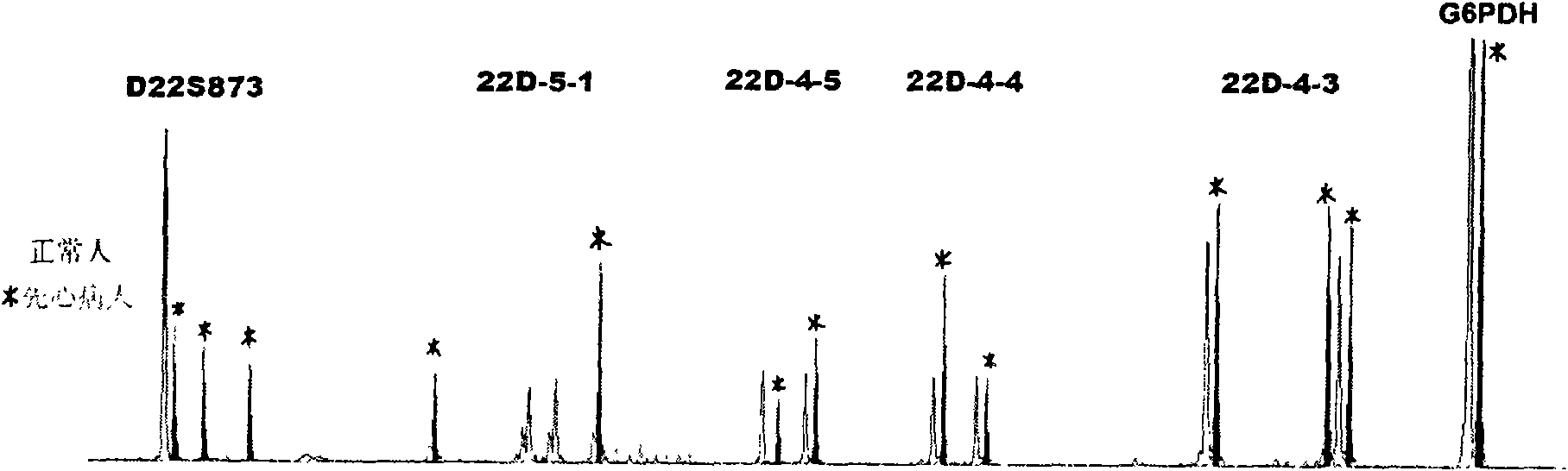
Method for testing chromosome 22q11.2 microdeletion and microduplication
An assay method and microdeletion technology, applied in the field of bioengineering, can solve the problems of unsatisfactory positive rate, unavoidable false positive rate, and difficulty in large-scale promotion, and achieve a high-cost and low-cost solution that is conducive to large-scale clinical use. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Steps:
[0049] 1. Genomic DNA extraction: Resin TM Genomic DNA Extraction Kit (Shanghai Saibaisheng Technology Co., Ltd.); congenital heart disease patients and normal blood; Genomic DNA was extracted separately according to the requirements of the kit.
[0050] 2. Multiplex fluorescent quantitative PCR compound amplification
[0051] The complex amplification system includes 5 STR markers: D22S873, 22D_5_1, 22D_4_5, 22D_4_4, 22D_4_3 and an internal reference G6PDH;
[0052] The primer sequence is shown in the table below (FAM fluorescent label is added to the forward primer F of each STR site)
[0053]
[0054]
[0055] PCR amplification:
[0056] 25μl system includes: 35-80ng genomic DNA
[0057] 2.5μl 10×STR buffer (buffer) (promega)
[0058] 0.1 μM D22S873 primer
[0059] 0.2 μM 22D_5_1 primer
[0060] 0.4 μM 22D_4_5 primer
[0061] 1.1 μM 22D_4_4 primer
[0062] 0...
Embodiment 2
[0084] Steps:
[0085] 1. Genomic DNA extraction: Resin-type TM Genomic DNA Extraction Kit (Shanghai Saibaisheng Technology Co., Ltd.); another congenital heart disease patient and normal blood; Genomic DNA was extracted separately according to the requirements of the kit.
[0086] 2. Multiplex fluorescent quantitative PCR compound amplification
[0087] The complex amplification system includes 5 STR markers: D22S873, 22D_5_1, 22D_4_5, 22D_4_4, 22D_4_3 and an internal reference G6PDH;
[0088] The primer sequence is shown in the table below (FAM fluorescent label is added to the forward primer F of each STR site)
[0089]
[0090]
[0091] PCR amplification:
[0092] 25μl system includes: 35-80ng genomic DNA
[0093] 2.5μl 10×STR buffer (promega)
[0094] 0.1 μM D22S873 primer
[0095] 0.2 μM 22D_5_1 primer
[0096] 0.4 μM 22D_4_5 primer
[0097] 1.1 μM 22D_4_4 primer
[0098] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com