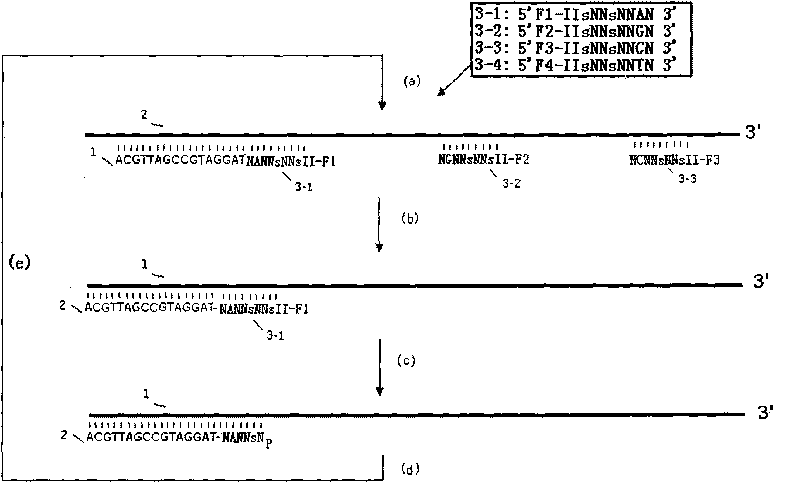
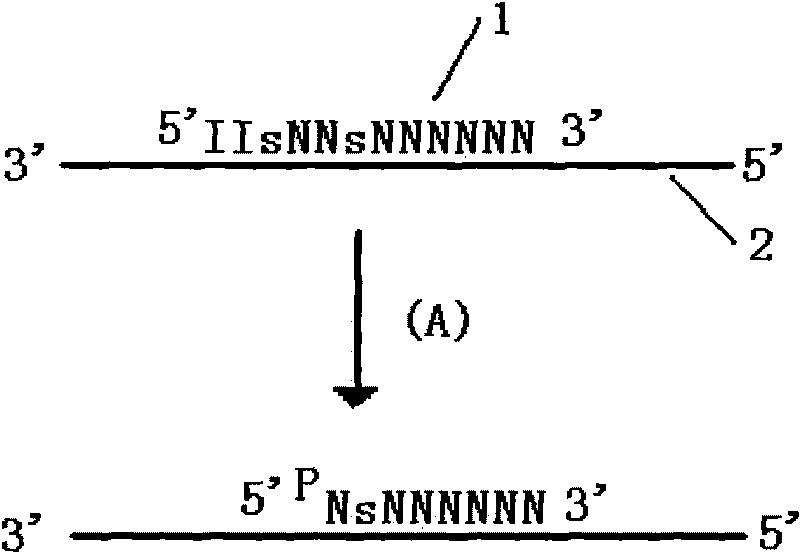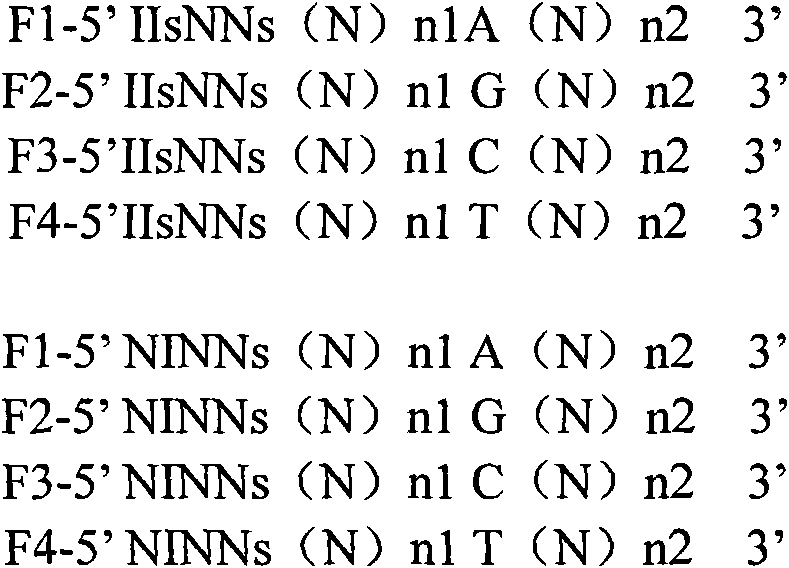Method for improving specificity in cutting position of endonuclease V
An endonuclease and specific technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as error and sequencing speed limit, achieve high cutting efficiency, increase read length, and cut position accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0033] Example 1: Hybridization-ligation Fluorescently Labeled Sequencing Determination of Inclusion of the Whole Human Genome.
[0034] Cut the human genome with enzymes (or sonicate) into fragments with a size of 50-1000 bases, and connect these fragmented nucleic acid sequences with a pair of universal linkers (assumed to be 20 bases in length) under the action of ligase Base), the oligonucleotide sequence of one universal linker is completely complementary to the sequence of the amplification primer, and the oligonucleotide sequence of the other linker is the same as that of the sequencing positioning primer.
[0035] The fragmented nucleic acid sequence connected by these tethers and the complementary sequence of the fixed linker are transferred to microbeads for emulsion parallel PCR reaction to amplify the fragmented whole human genome. These microbeads are immobilized on the plate substrate, and the human whole genome sequencing template is obtained by enzyme digestion...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com