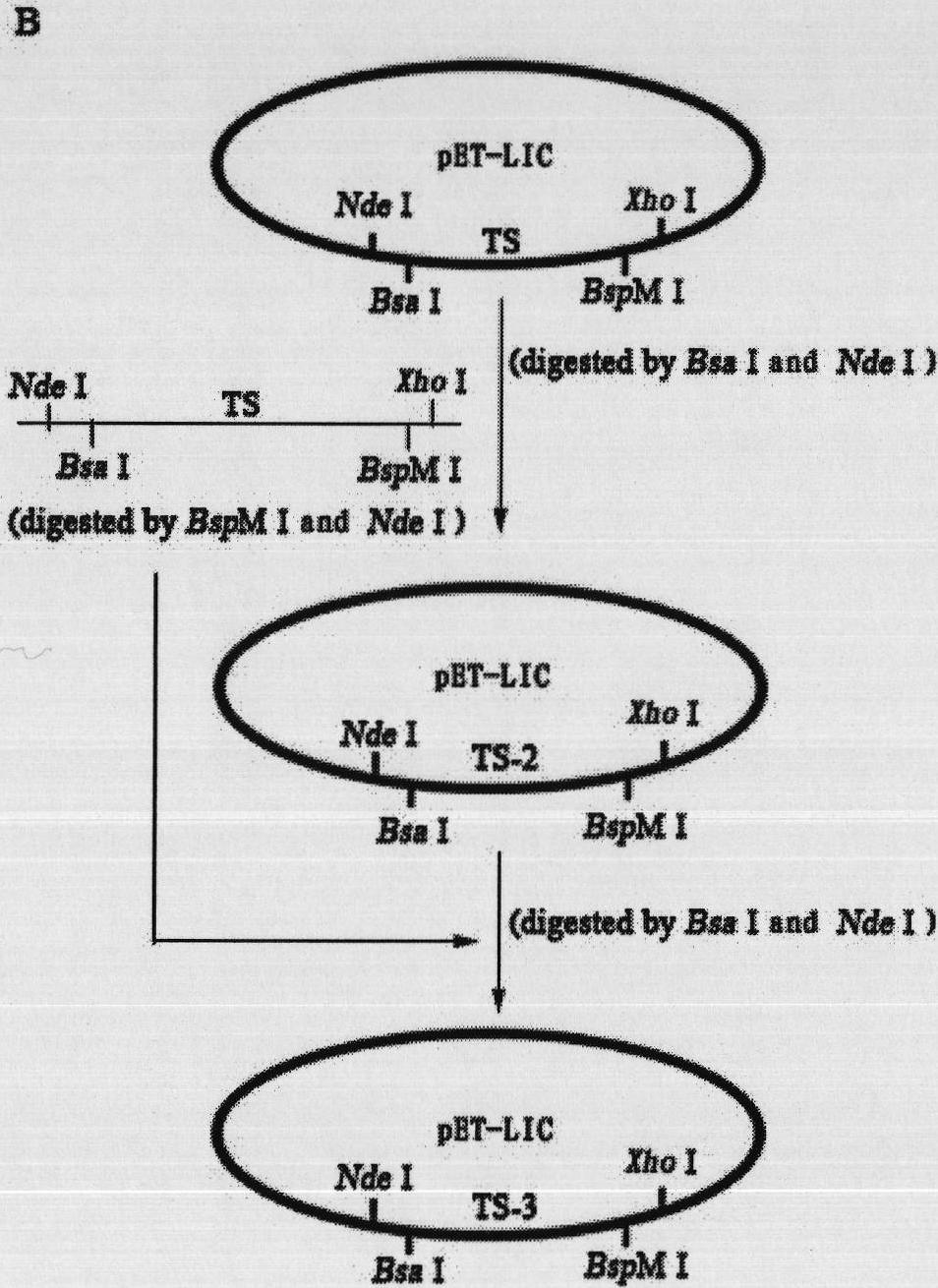
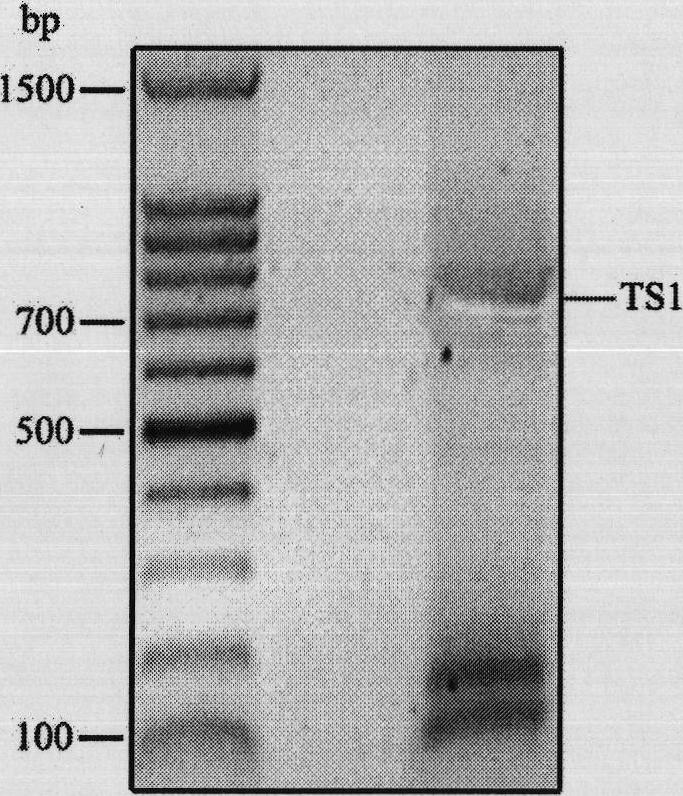
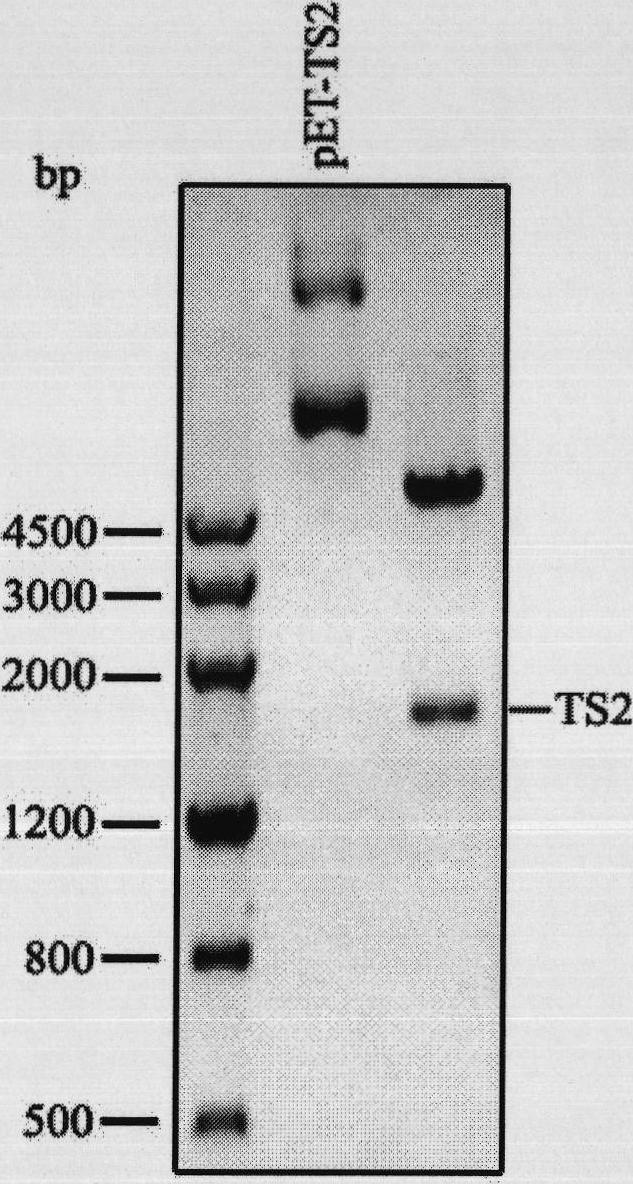
Repeated module gene-splicing method
A gene splicing and gene technology, applied in the field of repetitive module gene splicing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Take the spliced flagellar silk gene repeat framework as an example;
[0073] (1) Amplify the target gene by using the following PCR primers for the 696bp target repetitive gene;
[0074] Upstream primers:
[0075] 5'-ggt CATATG ttgg GGTCTC g
[0076] Nde I Bsa I
[0077] Downstream primers:
[0078] 5'-ggt CTCGAG ccaa ACCTGC TTCCAG 3'
[0079] Xho I BspM I
[0080] Among them, the underlined part is the enzyme cutting site, and the shaded part is the sequence matching part;
[0081] The 696bp gene sequence of the spider silk flagellar order:
[0082] CTGGTG CTGGTGTTTC TGGCGGTGTT
[0083] GGCCCTGGTG GCCTGGGTGG CCCTGGTGGT TTTGGCGGTC CGGGTGGCCC AGGTGGCCCT
[0084] GGTGGCCCGG GTGCACCTGG TGGCGAAGCG GGTGGTCTTT ATGGTCCGGG CGGTGCGGGT
[0085] GGCCTGTATG GCCCTGGCGG CGCGGGCGGT CTGTATGGTC CAGGCGGTGC GGGTGCACCG
[0086] GGTGGTCCAG GTGGTCCTGG TGGCTTTGGT GGCCCTGGCG GCCTGGGTGG TCCAGGTGGC
[0087] TTTGGCGGTG CAAGCGGTGC AGGCGCCGGT GGCGTTGGTC CGGGT...
Embodiment 2
[0112] Adding a repeat module to the 3' end of the silk repeat gene
[0113] The 819bp repeated spidroin gene was amplified by PCR;
[0114] Upstream primers:
[0115] 5'-ggc catatg aacc ggtctct -3';
[0116] Nde I Bsa I
[0117] Downstream primers:
[0118] 5'-ggc ctcgagc caa acctgcg ctcGGCG
[0119] Xho I BspM I
[0120] Among them, the underlined part is the enzyme cutting site, and the shaded part is the sequence matching part;
[0121] 819bp repeat spidroin protein target gene, the sequence is as follows:
[0122] CTGGGGTCAACGTGGTCCTCGCTCTCAAGGTCCTGGTTCTGGCGGTCAGCAGGGTCCGGGTGGTC
[0123] AGGGTCCTTATGGTCCTAGCGCGGCTGCAGCAGC
[0124] GGTCTGTCTCTGGAAGCAAAAACTAACGCTATC
[0125] GCTTCCGCACTGAGCGCGGCCTTCCTGGAAACCACGGGTTACGTTAATCA
[0126] GCAGTTTGTAAACGAAATCAAAACGCTGATCTTTATGATCGCACAGGCTT
[0127] CCTCCAACGAAATCTCTGGTTCTGCTGCAGCTGCGGGTGGTTCCAGCGGC
[0128] GGTGGTGGTGGCTCCGGTCAGGGTGGCTATGGTCAGGGCGCCTACGCCTC
[0129] TGCCAGCGCGGCGGCTGCCTA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com