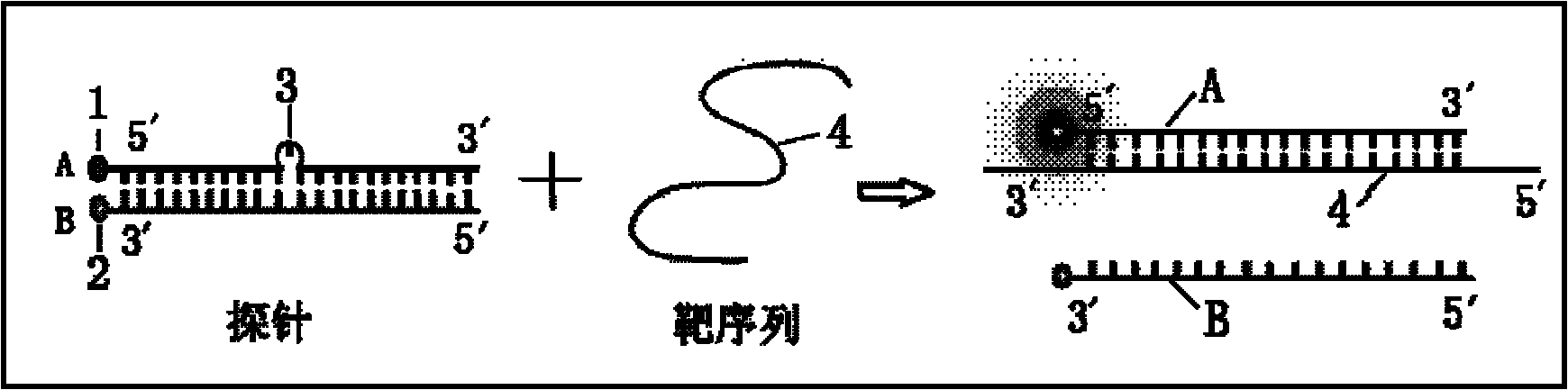
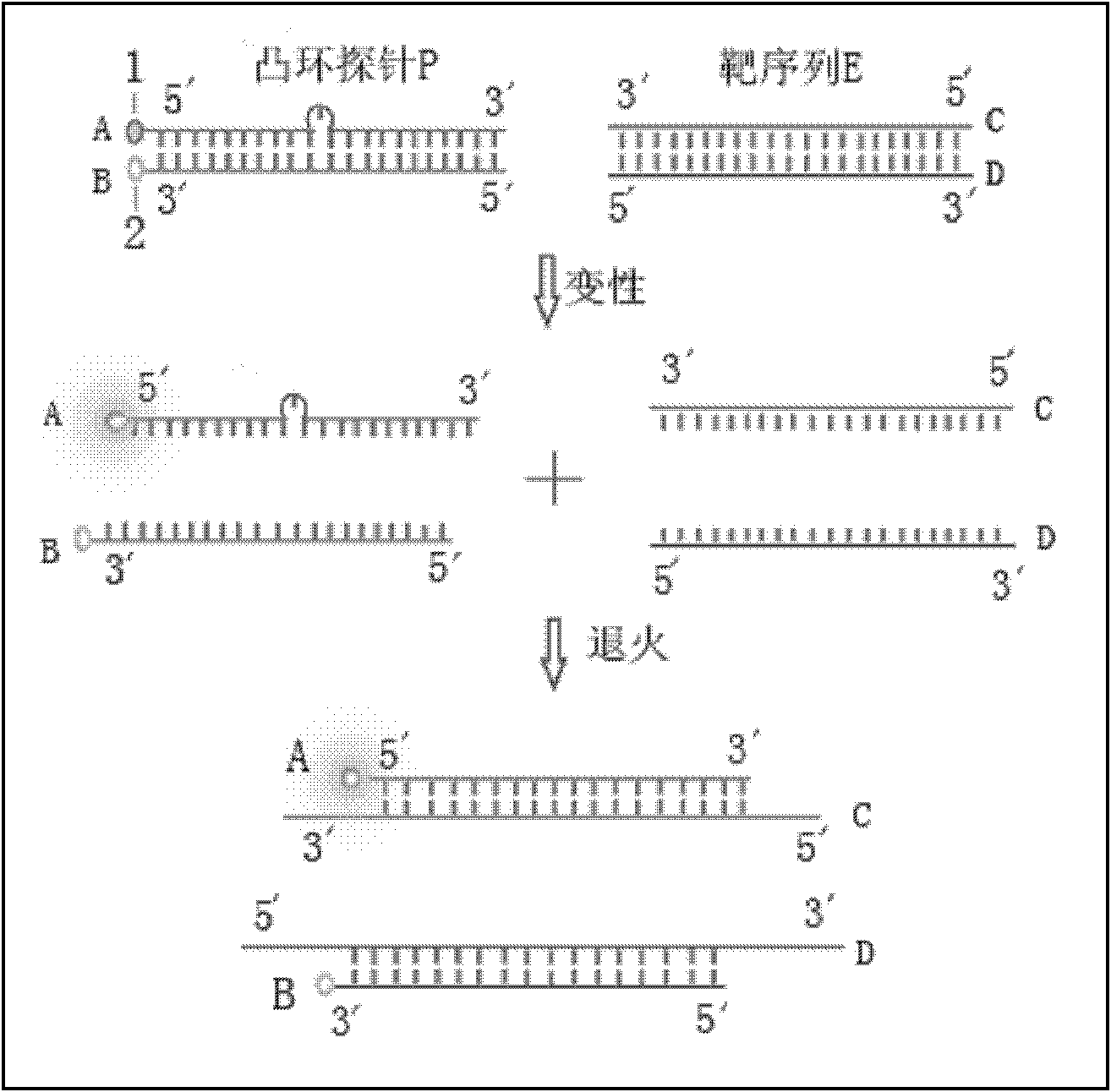Probe used for identifying and detecting nucleic acid specificity
A specific recognition and probe technology, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, fluorescence/phosphorescence, etc., can solve the problems of high fluorescence background, limited single-base mutation recognition ability, complex design, etc., to achieve The effect of high hybridization efficiency, strong tolerance, and simple design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1: Real-time PCR detects hepatitis B virus
[0042] In order to investigate the effectiveness of the protruding ring probe in real-time PCR detection, the PCR template is a series of diluted standard DNA samples. The total volume of the reaction system is 50 μl, including 5 μl 10×buffer (100 mM [NH 4 ] 2 SO 4 , 6700mM Tris-HCl pH 8.3), 1.5μl 25mM MgCl 2 , each dNTP 400 μM, upstream and downstream primers 0.4 μM, probe 0.1 μM, 2.0U Taq DNA polymerase, 5 μl template DNA. The real-time PCR reaction was carried out on the MX3000P real-time PCR instrument. The reaction conditions were: pre-denaturation at 94°C for 3 minutes, followed by 25s at 94°C, 20s at 58°C (detection of FAM and fluorescence signal), 15s at 72°C, a total of 35 cycles. Serial 10-fold serial dilution of standard DNA, H 2 O served as a negative control. The convex loop probe includes a nucleic acid sequence that is complementary to the amplified product. The upstream and downstream primer...
Embodiment 2
[0045] Example 2: Application of convex ring probes in the detection of SNPs
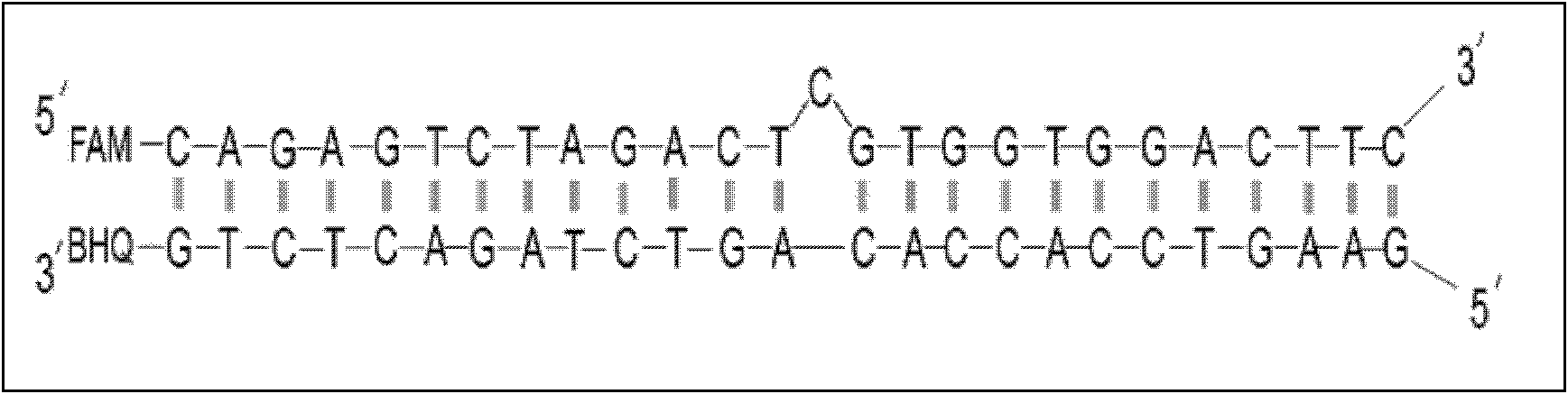
[0046] Select the HPA-4 gene of human platelet alloantigen (human platelet alloantigen, HPA), because there is a G>A mutation in the gene, resulting in the production of two platelet antigens, HPA-4a and HPA-4b, HPA-4a It is the wild type, and HPA-4b is the mutant type. Protrusion loop probes were designed for the gene sequences of the two antigens, and the difference between the two probes was only 1 base, and 3 typical samples with known genotypes were selected for testing. One is a pure and HPA-4a sample, one is a pure and HPA-4b sample, and one is a heterozygous sample containing both HPA-4a and HPA-4b. At the same time as the test, do a no (negative) sample, that is, H 2 O control. The first strand sequence of the wild-type (HPA-4a) probe is:
[0047] 5'-FAM-GGTGAGCTTT C GCATCTGGGT-3', the second strand sequence is 5'
[0048] -ACCCAGATGCAAAGCTCACC-3'-BHQ, mutant (HPA-4b) probe sequence i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com