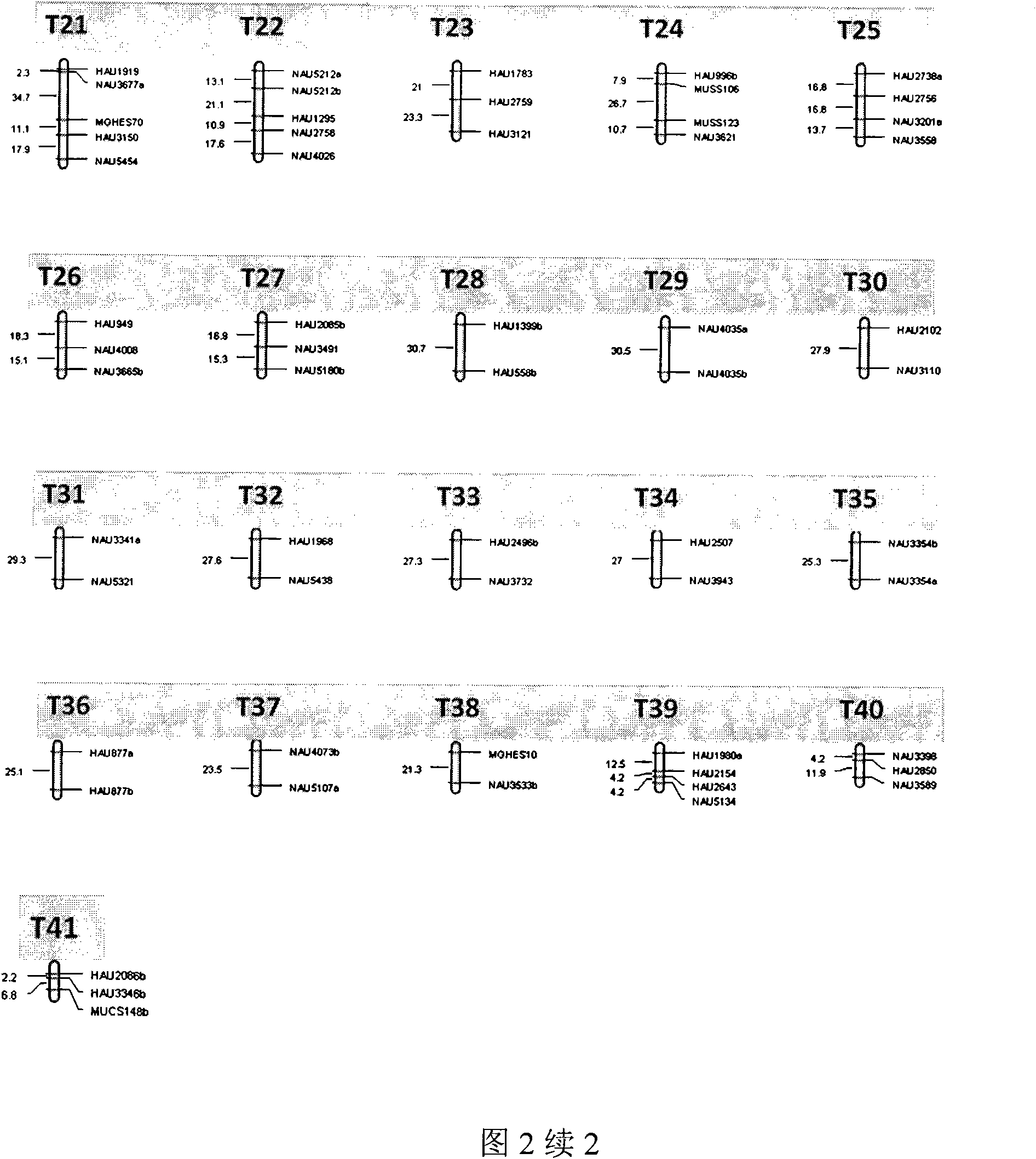Method for building cotton fiber transcription genetic linkage map by EST-SSR sign
A cotton fiber and map technology, applied in biochemical equipment and methods, microbe measurement/inspection, etc., can solve the problems of inability to study genetic information, complex RNA extraction, high cost, etc., and achieve the effect of fast and simple methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
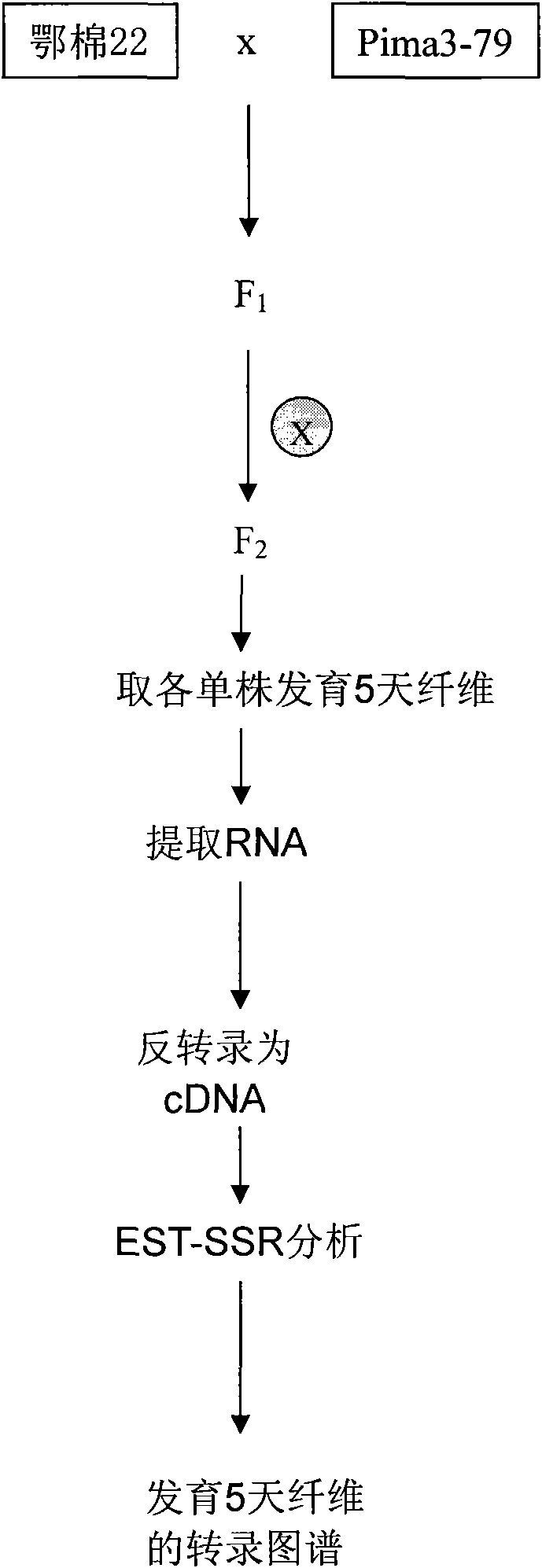
[0029] Sea-island cotton Pima 3-79 was used as the male parent (Li Wu et al., SRAP marker analysis of sea-island cotton genetic diversity, ACTAAGRONOMICA SINICA 2008, 34(5): 893-898), upland cotton variety Emian 22 (ZhangYanxin et al. ,
[0030] Characteristics and analysis of simple sequence repeats in the cotton genome based on linkage map constructed from a BC 1 Population between gossypium hirsutum and G.barbadense, Genome, 2008(51) as the female parent, obtained F1, planted F1 in Hainan breeding base, China, self-crossed F1, obtained F2, planted F2 in Wuchang Huazhong Agriculture, Wuhan City, Hubei Province University experiment Tanaka, using this F2 single plant as the starting material for mapping the fiber transcription map;
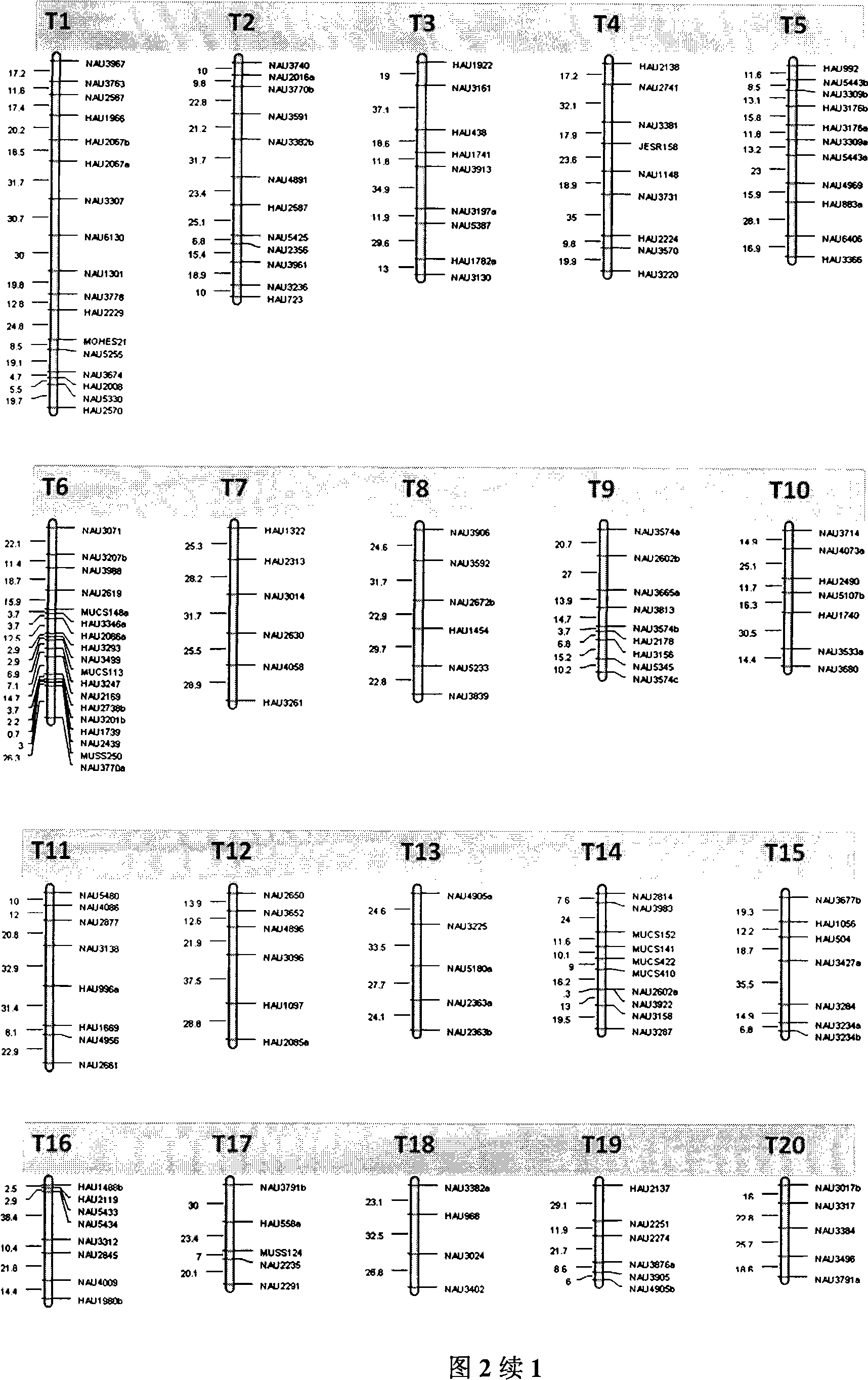
[0031] 2. Construct transcriptional map from the developing cotton fiber 5 days after anthesis. Specific steps are as follows:
[0032] Material preparation
[0033] Cotton fibers developed for 5 days were taken. Every morning in the field,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com