Rice blast resistance gene Pi1 and application thereof
A resistance gene and rice blast resistance technology, applied in the field of genetic engineering, can solve problems such as the lack of effective resistance genes, unresolved problems, and the susceptibility of disease-resistant varieties, and achieve the effect of shortening the breeding time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Resistance characteristics of resistance gene Pi1
[0038] First, in order to compare and clarify the resistance spectrum of the five alleles (Pi1, Pik-p, Pik, Pik-m, pi1) at the Pi1 locus, using 40), Hunan (HN, 40), Yunnan (YN, 43), Guizhou (GZ, 60), Sichuan (SC, 66), Jiangsu (JS, 72), Liaoning (LN, 108 ), Jilin (JL, 60), Heilongjiang (HLJ, 63), a total of 612 strains collected were resistant to the varieties held by the above five alleles (respectively C101LAC, K60, Kusabue, Tsuyuake and CO39) comparative analysis. The results showed that Pi1 showed high resistance to 8 rice blast fungus populations except Liaoning and Jilin. This shows that the gene can be used in the above regions. Rice varieties C101LAC and CO39 have been disclosed in the literature (Inukai et al. Allelism of blastresistance genes in near-isogenic lines of rice. Phytopathology, 1994, 84: 1278-1283). Other rice varieties have been disclosed in the literature (Wang et al. Characterizati...
Embodiment 2
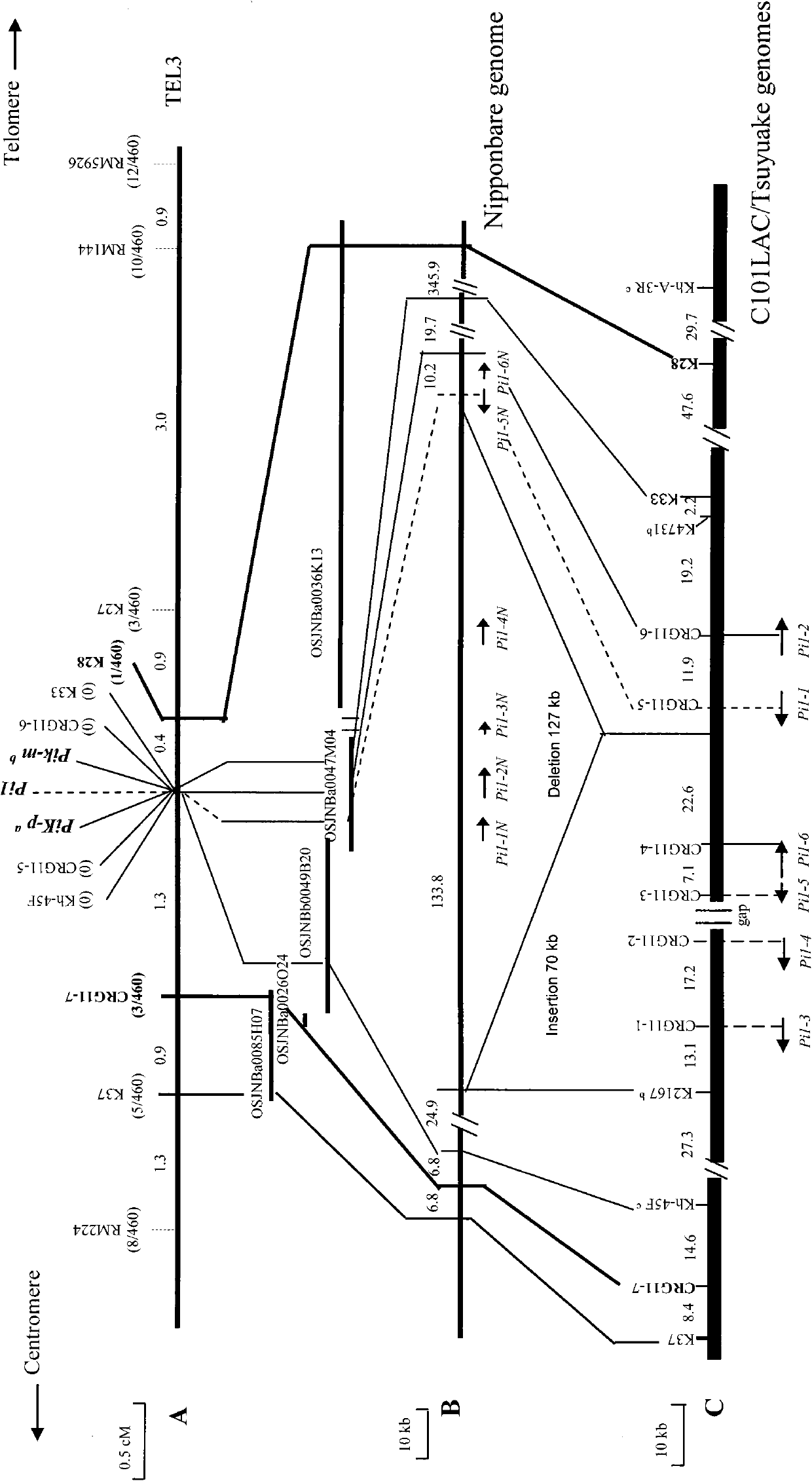
[0039] Example 2 Location and Electron Physical Mapping of Rice Blast Resistance Gene Pi1
[0040] F 2 Population, inoculated with the strain CHL930 (Wang et al. Characterization of rice blast resistance genes in the Pik cluster and fine mapping of the Pik-p locus. Phytopathology, 2009, 99 :900-905). The results showed that the F 2 The segregation ratio of resistant plants and susceptible plants in the population was 3:1. It was inferred that the resistance of C101LAC was controlled by a pair of dominant genes. Therefore, a mapping population consisting of 230 individuals (equivalent to 460 gametophytes) was constructed.
[0041] The previous genetic analysis showed that the Pi1 gene and the Pik cluster gene are closely linked (Inukai et al.1994, Phytopathology, 84: 1278-1283), and the applicant's laboratory has carried out fine mapping of the Pik-m gene (Li et al. 2007, Molecular Breeding, 20: 179-188). Therefore, applicants performed linkage analysis using molecular ma...
Embodiment 3
[0043] Example 3 Annotation and sequence analysis of rice blast resistance gene Pi1 candidate gene
[0044] In order to determine the candidate gene of Pi1, the applicant used the reference sequence of Nipponbare, through three gene prediction software RiceGAAS (http: / / ricegaas.dna.affrc.go.jp), Gramene (http: / / 143.48.220.116 / resources / ) and Softberry's FGENESH (http: / / www.softberry.com) performed gene prediction and annotation analysis on the target gene region, and preliminarily determined that the candidate resistance gene of Pi1 is 6 nucleotide binding sites and leucine-rich Candidate genes (Pi1-1N, Pi1-2N, Pi1-3N, Pi1-4N, Pi1-5N, and Pi1-6N) of nucleotide binding site-leucine-rich repeat (NBS-LRR).
[0045] The presence / absence (P / A) analysis of 6 candidate genes based on gene-specific markers showed that the 4 candidate genes (Pi1-1N, Pi1-2N, Pi1- 3N and Pi1-4N) were absent in C101LAC. According to the information provided by Ashikawa et al. (2008, Genetics, 180: 2267...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com