Multicolor fluorescence composite detection method and kit of 44 SNPs (Single Nucleotide Polymorphism) loci
A composite detection and kit technology, applied in the field of forensic individual identification, can solve problems such as unsuitable for micro-quantity test materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
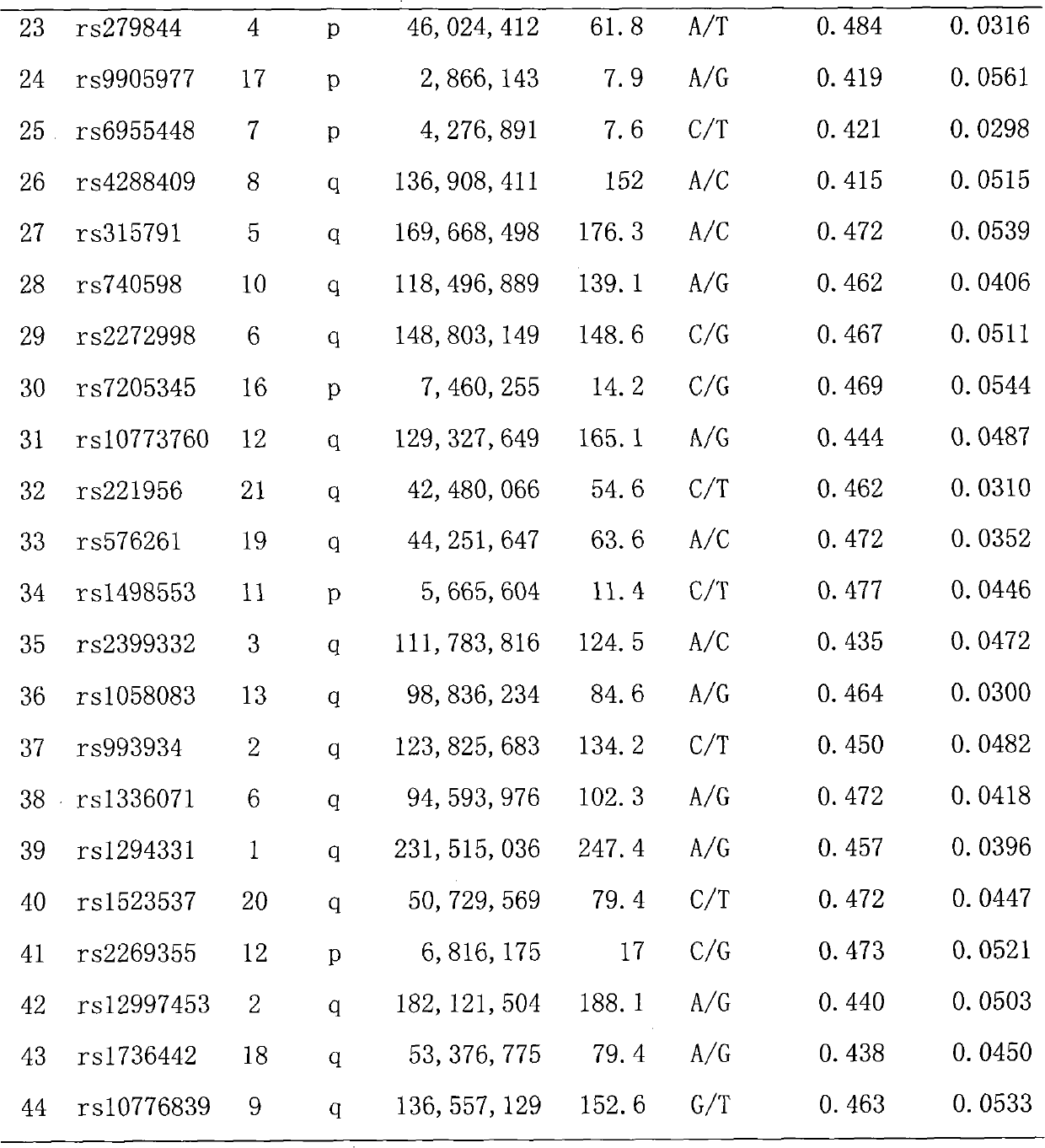
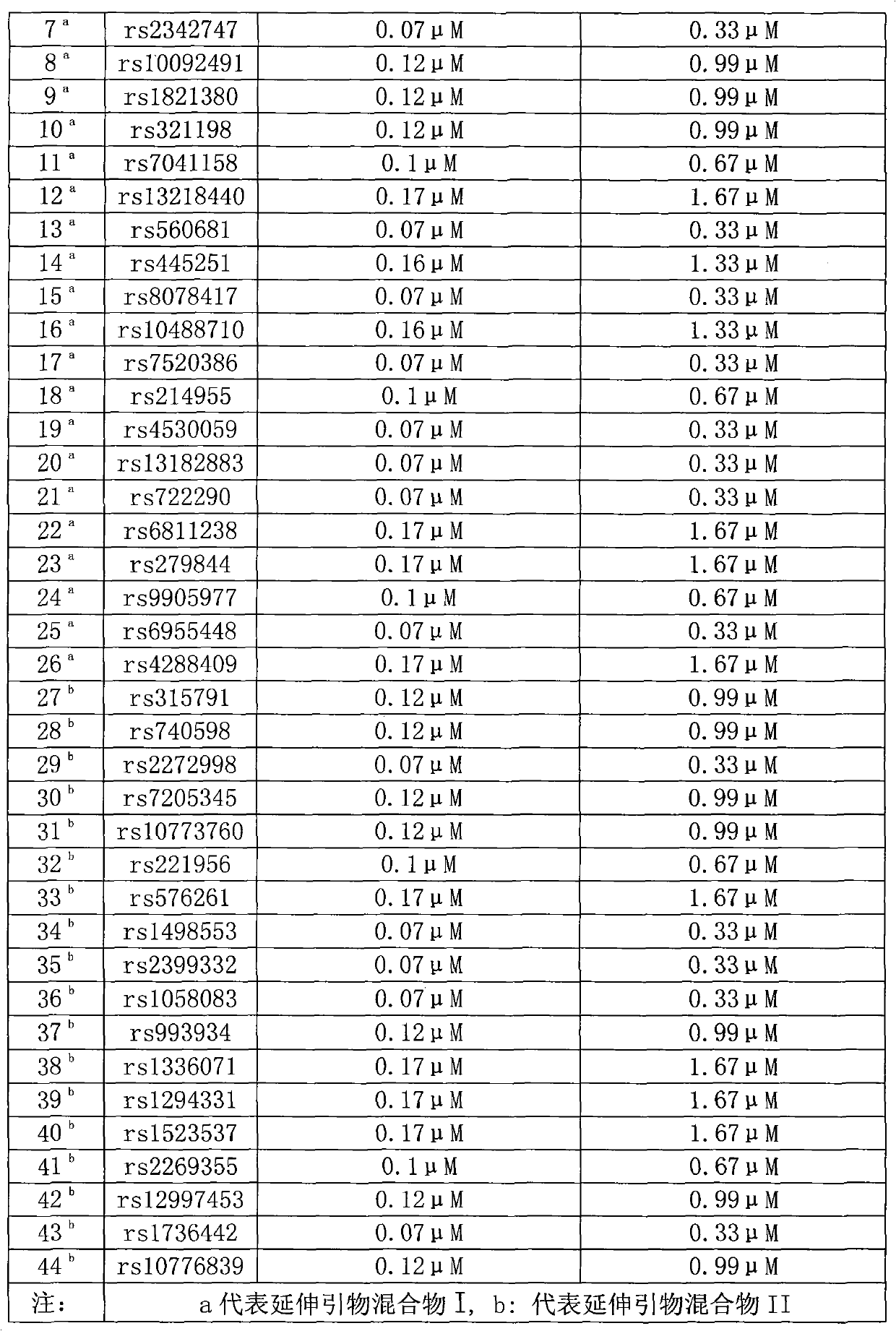
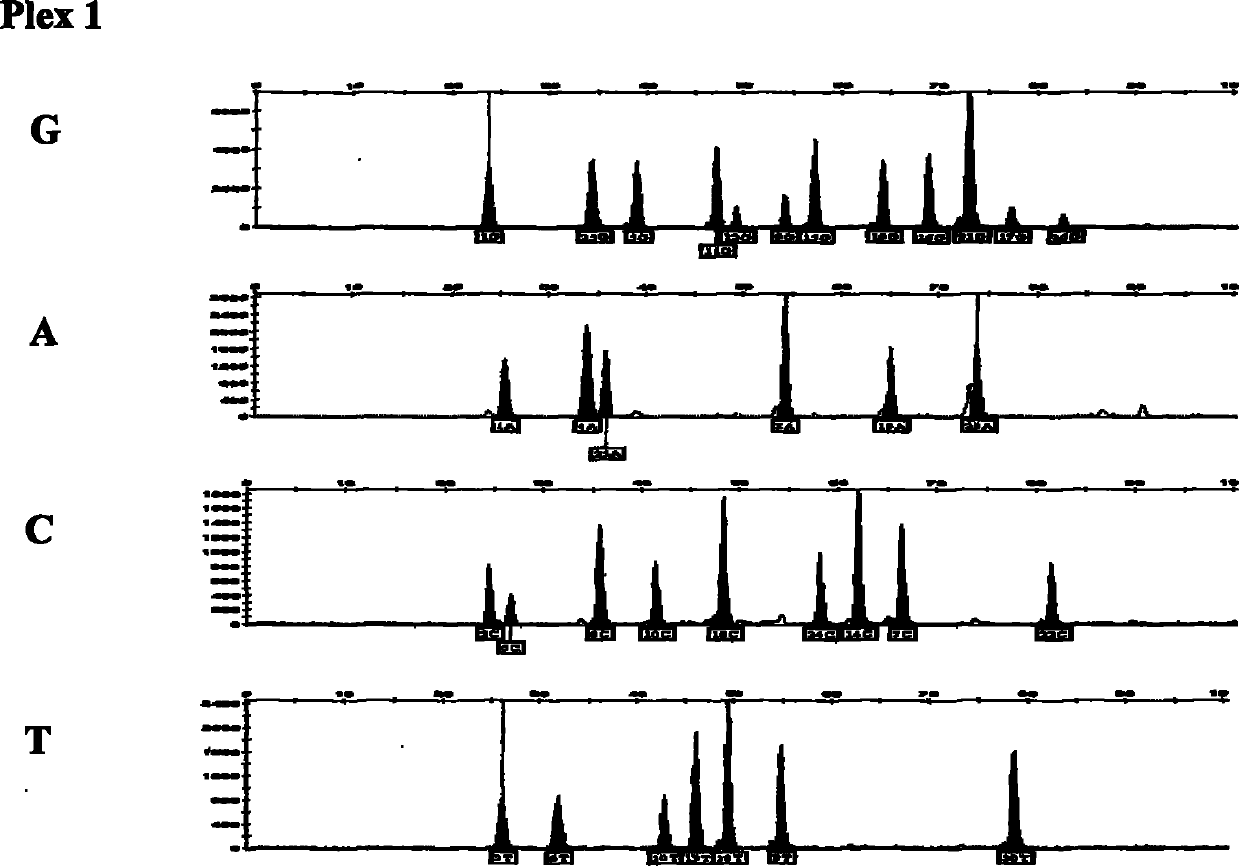
[0027] 44 SNP site multi-color fluorescent composite detection kits for individual identification, including a) amplification system: PCR buffer solution, MgCl 2 , dNTPs, PCR primers, Taq DNA polymerase, template DNA, b) amplification product purification reagents: exonuclease Ⅰ and its buffer solution, shrimp alkaline phosphatase and its buffer solution, c) extension reaction reagent: extension primer and SNaPshot reaction mixture, d) Test reagents: Hi-Di formamide and GeneScan TMSize Standards-120LIZ, the PCR primers and extension primers are composed of primers for 44 SNP gene loci, and the rs numbers of the 44 SNP gene loci in the SNP database in NCBI are rs1109037, rs3780962, rs987640, rs9951171, rs430046 respectively ,rs338882,rs2342747,rs10092491,rs1821380,rs321198,rs7041158,rs13218440,rs560681,rs445251,rs8078417,rs10488710,rs7520386,rs214955,rs4530059,rs13182883,rs722290,rs6811238,rs279844,rs9905977,rs6955448,rs4288409,rs315791,rs740598,rs2272998,rs7205345 , rs1077376...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com