Method for evaluating anti-cancer drug inhibiting aggregation of membrane protein receptors
A technology of anti-cancer drug and identification method, which is applied in the field of anti-cancer drug identification, can solve the problems of long cycle, high cost, difficult drug screening, etc., and achieve the effect of short cycle, easy operation, and cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032]1) Transfer the prepared HeLa cells to a 35mm glass bottom culture dish and incubate at 37°C for 24 hours to make the cell coverage reach 80%, and then transfect the TβRII-GFP plasmid into this culture dish to make the final concentration Incubate at 37°C for 5 hours in serum-free DMEM at 1 μg / ml to express TβRII-GFP protein on HeLa cells to obtain recombinant cancer cells, then wash the culture dish with serum-free DMEM for 3-5 times.
[0033] The TβRII-GFP plasmid used (Single-molecule imaging reveals transforming growth factor-β-induced type II receptor dimerization; Wei Zhanga, Yaxin Jianga, Qiang Wangb, Xinyong Maa, Zeyu Xiaoa, Wei Zuob, Xiaohong Fanga, 1, and Ye-Guang Chenb, PNAS, September 15, 2009, vol.106, no.37, 15679-15683), the plasmid can be obtained from the Institute of Chemistry, Chinese Academy of Sciences.
[0034] 2) Add naringenin to the cell culture dish used in step 1) to a final concentration of 50 μM, and incubate in serum-free DMEM at 37° C. for ...
Embodiment 2
[0049] According to the same method and steps as in Example 1, the drug to be identified was identified, only the Hela cells used were replaced with MCF7 cells.
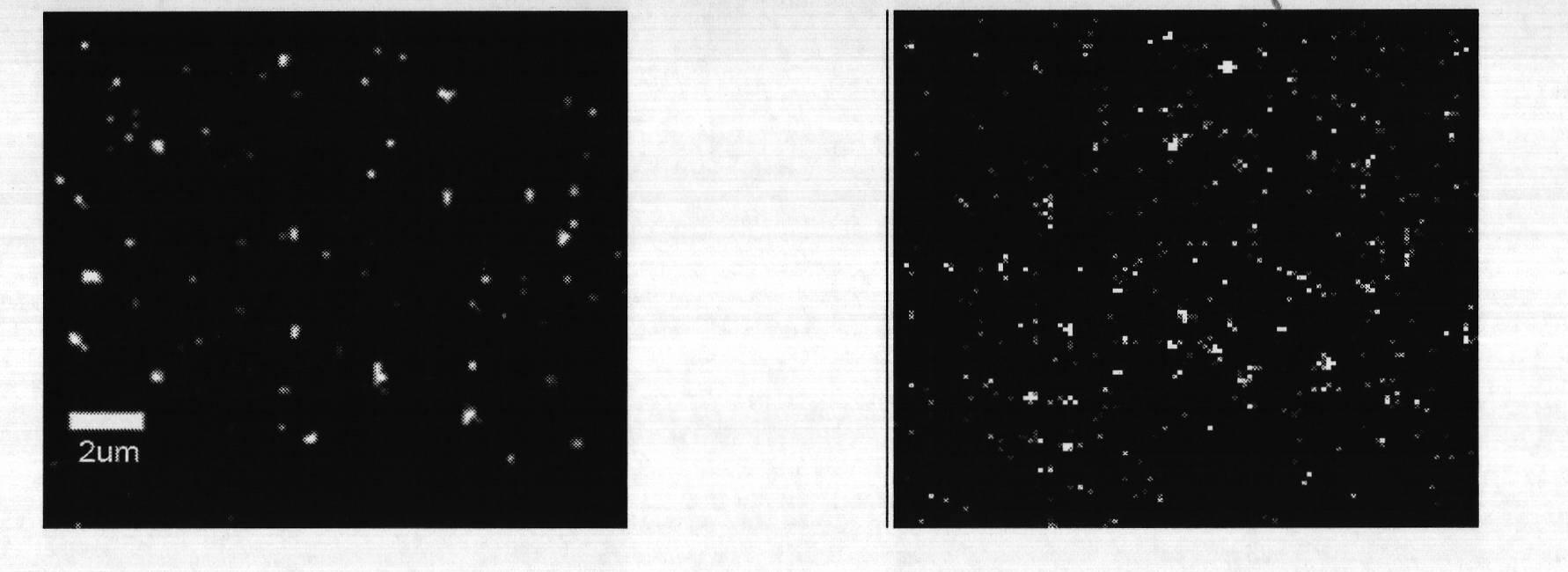
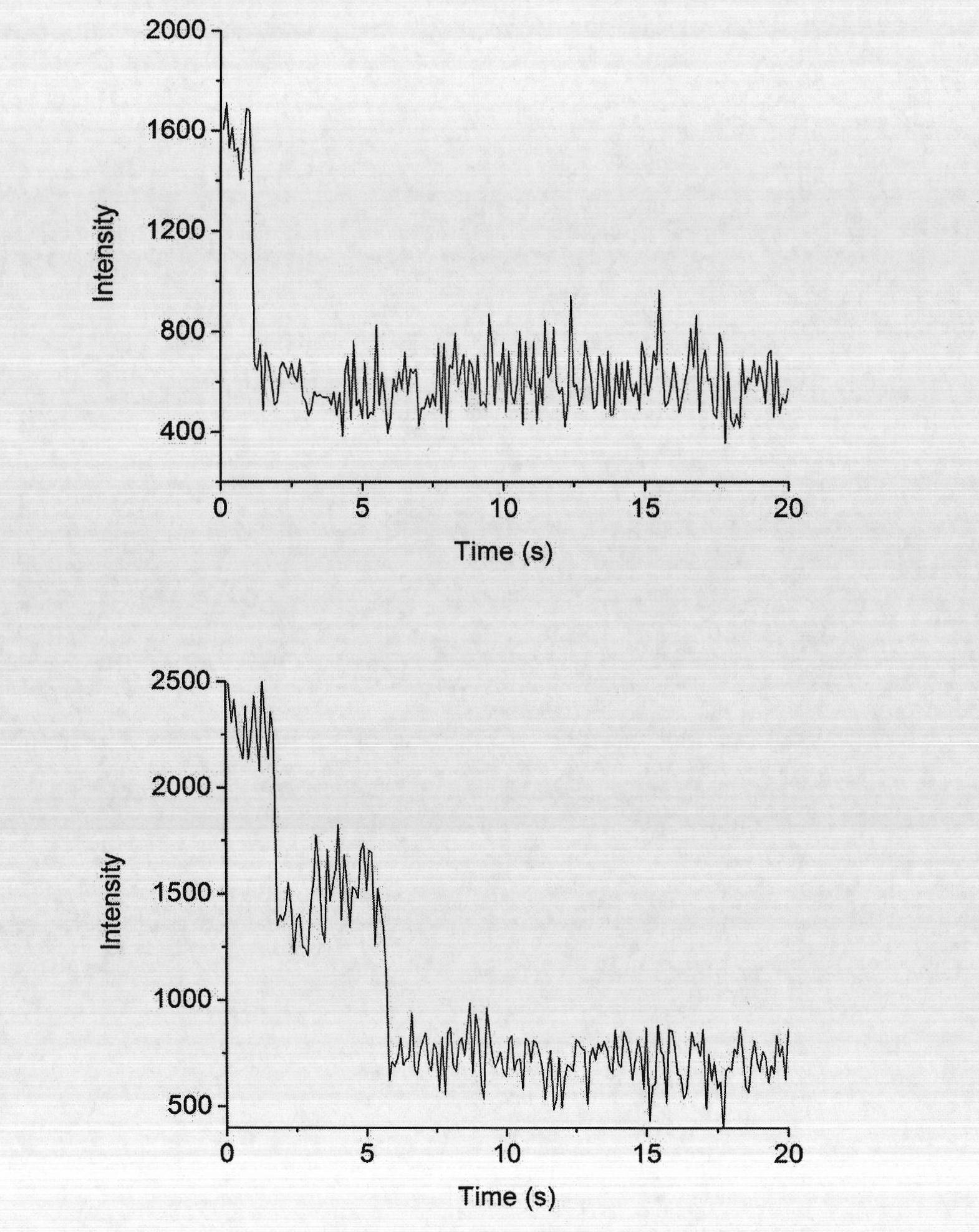
[0050] Fluorescent bright spots less than or equal to 5×5 pixels in the cell images were counted. if so figure 2 The one-step bleaching curve shown on the left is TβRII-GFP monomer. if so figure 2 The two-step bleaching curve shown on the right is TβRII-GFP dimer.
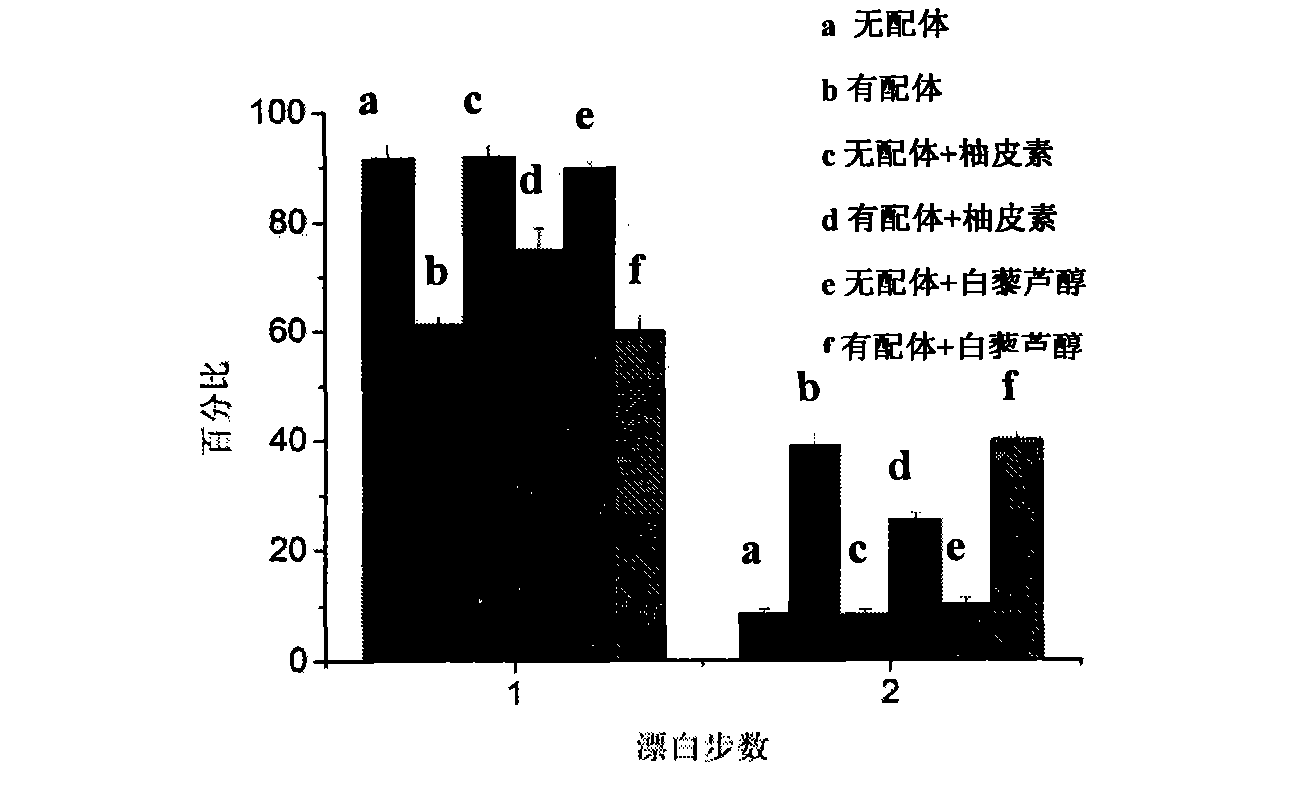
[0051] The results are as follows: the statistics of the bleaching steps of a single fluorescent point are as follows: Figure 4 As shown, when the drug to be identified was not added, after adding the ligand, TβRII-GFP aggregated, and the ratio of TβRII-GFP dimer in the bleaching step A was 37.7±2.9%. After adding 50μM naringenin and incubating for 1h, the proportion of TβRII-GFP aggregation in bleaching step B was significantly reduced, and the proportion of TβRII-GFP dimer decreased to 26.3±1.8%.
[0052] Biochemical experiment verification:
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| refractive index | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com