Method for assisting in identifying root-knot nematodes and special primer pair thereof
A technology of root knot nematode and southern root knot nematode, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragment, etc., can solve problems such as unreachable, low SCAR sensitivity, and long time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, the preparation of primer
[0035] Artificially synthesized the following four primers:
[0036] #C2F3: 5'-GGTCAA TGTCAGAAA TTTGTGG-3'; Sequence 1;
[0037] #1108: 5'-TACCTTTGACCAA TCACGCT-3'; Sequence 2;
[0038] MI-F: 5'-GGGCAAGTAAGGATGCTCTGAC-3'; Sequence 3;
[0039] MI-R (5'-CTTTCATAGCCACGTCGCGATC-3'; SEQ ID NO: 4.
[0040] The primer pair consisting of #C2F3 and #1108 was used as primer pair A, and the primer pair consisting of MI-F and MI-R was used as primer pair B. Primer pair A targets the region between CO II and LrRNA in mitochondrial DNA (mtDNA). Primer pair B is directed against the esophageal adenin gene.
Embodiment 2
[0041] Embodiment 2, application primer is assisted in distinguishing root-knot nematode (adult)
[0042] 1. Using primers to help identify root-knot nematodes
[0043] 1. Extract the genomic DNA of a single root-knot nematode (adult).
[0044] 2. Using genomic DNA as a template to carry out PCR reaction to obtain PCR amplification products.
[0045] PCR reaction system (25 μL): 17.1 μl ddH20; 2.5 μl 10× buffer; 2 μl dNTP (2.5 mM); 1 μl primer #C2F3 (20 μM); 1 μl primer #1108 (20 μM); 0.4 μl Taq enzyme (5U / μl); 1 μl template.
[0046] PCR reaction program: pre-denaturation at 94°C for 4 minutes; 40 cycles of denaturation at 94°C for 1 minute, annealing at 48°C for 1 minute, and extension at 72°C for 2 minutes; finally, incubation at 72°C for 5 minutes.
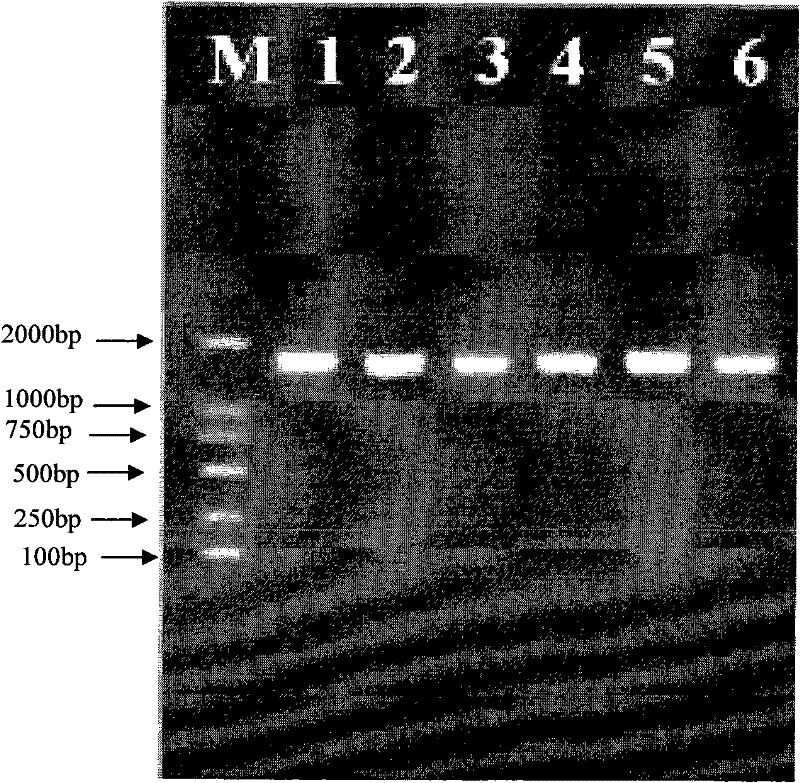
[0047] 3. The PCR amplification product was subjected to 1% (containing 0.5 μg / mL EB) agarose gel electrophoresis, the electrophoresis buffer was 0.5×TBE, the voltage was 80 V, and the electrophoresis was about 30 min. Pho...
Embodiment 3
[0056] Embodiment 3, application primer pair assists in identifying root-knot nematode larvae (2 instar larvae)
[0057] 1. Using primers to help identify root-knot nematodes
[0058] 1. Extract the genomic DNA of a single root-knot nematode (2nd instar larva).
[0059] 2. Using genomic DNA as a template to carry out PCR reaction to obtain PCR amplification products.
[0060] PCR reaction system (25 μL): 17.1 μl ddH20; 2.5 μl 10× buffer; 2 μl dNTP (2.5 mM); 1 μl primer #C2F3 (20 μM); 1 μl primer #1108 (20 μM); 0.4 μl Taq enzyme (5U / μl); 1 μl template.
[0061] PCR reaction program: pre-denaturation at 94°C for 4 minutes; 40 cycles of denaturation at 94°C for 1 minute, annealing at 48°C for 1 minute, and extension at 72°C for 2 minutes; finally, incubation at 72°C for 5 minutes.
[0062] 3. The PCR amplification product was subjected to 1% (containing 0.5 μg / mL EB) agarose gel electrophoresis, the electrophoresis buffer was 0.5×TBE, the voltage was 80 V, and the electrophore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com