Gene combination for instructing individualized treatment by medicines such as geftinat and Tarceva
A gene combination and drug technology, applied in the field of genes, can solve problems such as delaying the timing of treatment, failure to achieve treatment goals, and complicated procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 A method for identifying EGFR-Exon19 mutations
[0031] 1. Extract the genomic DNA of the patient's tumor tissue;
[0032] 2. Perform PCR amplification. The 20 μl PCR reaction system is as follows: 50-100 ng of genomic DNA of the tumor tissue to be tested, 0.125 μl of Taq enzyme, 1 μl of upstream and downstream primers (10 μM), 2 μl of dNTP (2.5 mM), 10 × PCR buffer ( Contains Mg 2+ ) 2 μl, and the rest was sterile distilled water; the PCR reaction conditions were: denaturation at 95°C for 2 min, followed by denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 1 min, and 45 cycles, and finally extension at 72°C for 7 min, and storage at 4°C;
[0033] 3. Gel electrophoresis analysis of PCR amplification products:
[0034] a. Rinse the electrophoresis tank and comb with distilled water. Put it on a level table and set up the comb. 1.5% agarose can be prepared for electrophoresis.
[0035] b. Put 50ml of 1×TAE electrophoresis ...
Embodiment 2
[0054] Example 2 A method for identifying EGFR-Exon21 mutations
[0055] 1. Extract the genomic DNA of the patient's tumor tissue;
[0056] 2. Perform PCR amplification. The 20 μl PCR reaction system is as follows: 50-100 ng of genomic DNA of the tumor tissue to be tested, 0.125 μl of Taq enzyme, 1 μl of upstream and downstream primers (10 μM), 2 μl of dNTP (2.5 mM), 10 × PCR buffer ( Contains Mg 2+ ) 2 μl, and the rest was sterile distilled water; the PCR reaction conditions were: denaturation at 95°C for 2 min, followed by denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 1 min, and 45 cycles, and finally extension at 72°C for 7 min, and storage at 4°C;
[0057] 3. Gel electrophoresis analysis of the PCR amplification product: the specific steps are the same as in Example 1.
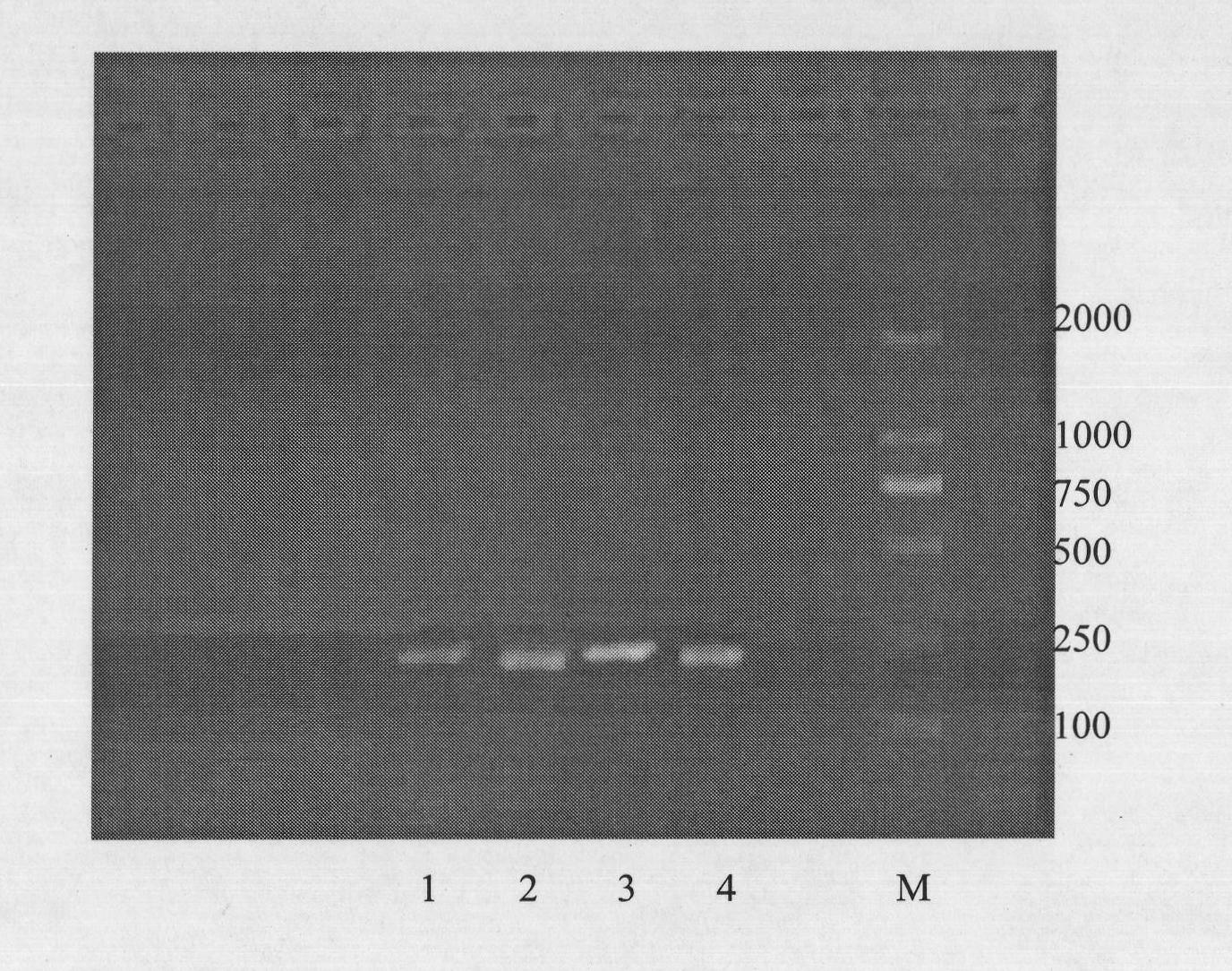
[0058] The result is as figure 1 As shown, band 4 represents EGFR-Exon21, and its PCR product is 247bp.
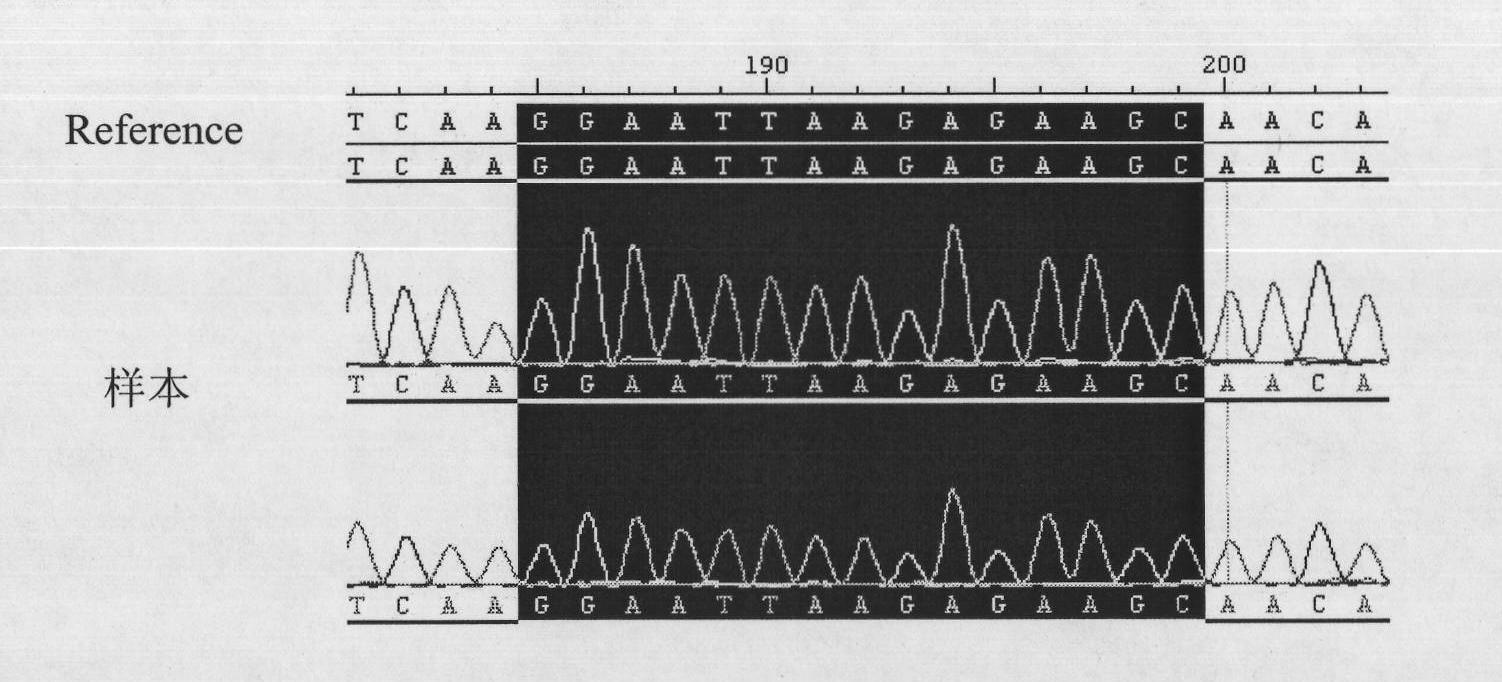
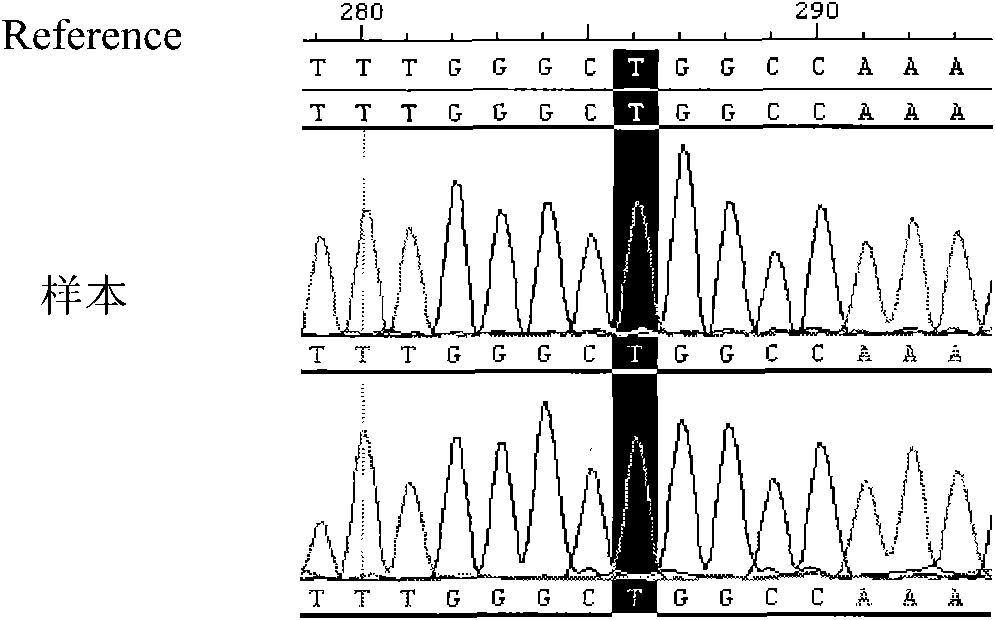
[0059] 4. Sequencing of PCR amplification products: the ...
Embodiment 3
[0061] Example 3 A method for identifying Kras-codon12 / 13 mutations
[0062] 1. Extract the genomic DNA of the patient's tumor tissue;
[0063] 2. Perform PCR amplification. The 20 μl PCR reaction system is as follows: 50-100 ng of genomic DNA of the tumor tissue to be tested, 0.125 μl of Taq enzyme, 1 μl of upstream and downstream primers (10 μM), 2 μl of dNTP (2.5 mM), 10 × PCR buffer ( Contains Mg 2+ ) 2 μl, the rest was sterile distilled water; the PCR reaction conditions were: denaturation at 95°C for 2 minutes, followed by denaturation at 95°C for 30 sec, annealing at 50°C for 30 sec, extension at 72°C for 1 min, and 45 cycles, and finally extension at 72°C for 7 min, and storage at 4°C;
[0064] 3. Gel electrophoresis analysis of the PCR amplification product: the specific steps are the same as in Example 1.
[0065] The result is as figure 1 As shown, band 1 represents Kras-codon12 / 13, and its PCR product is 235bp.
[0066] 4. Sequencing of PCR amplification produc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com