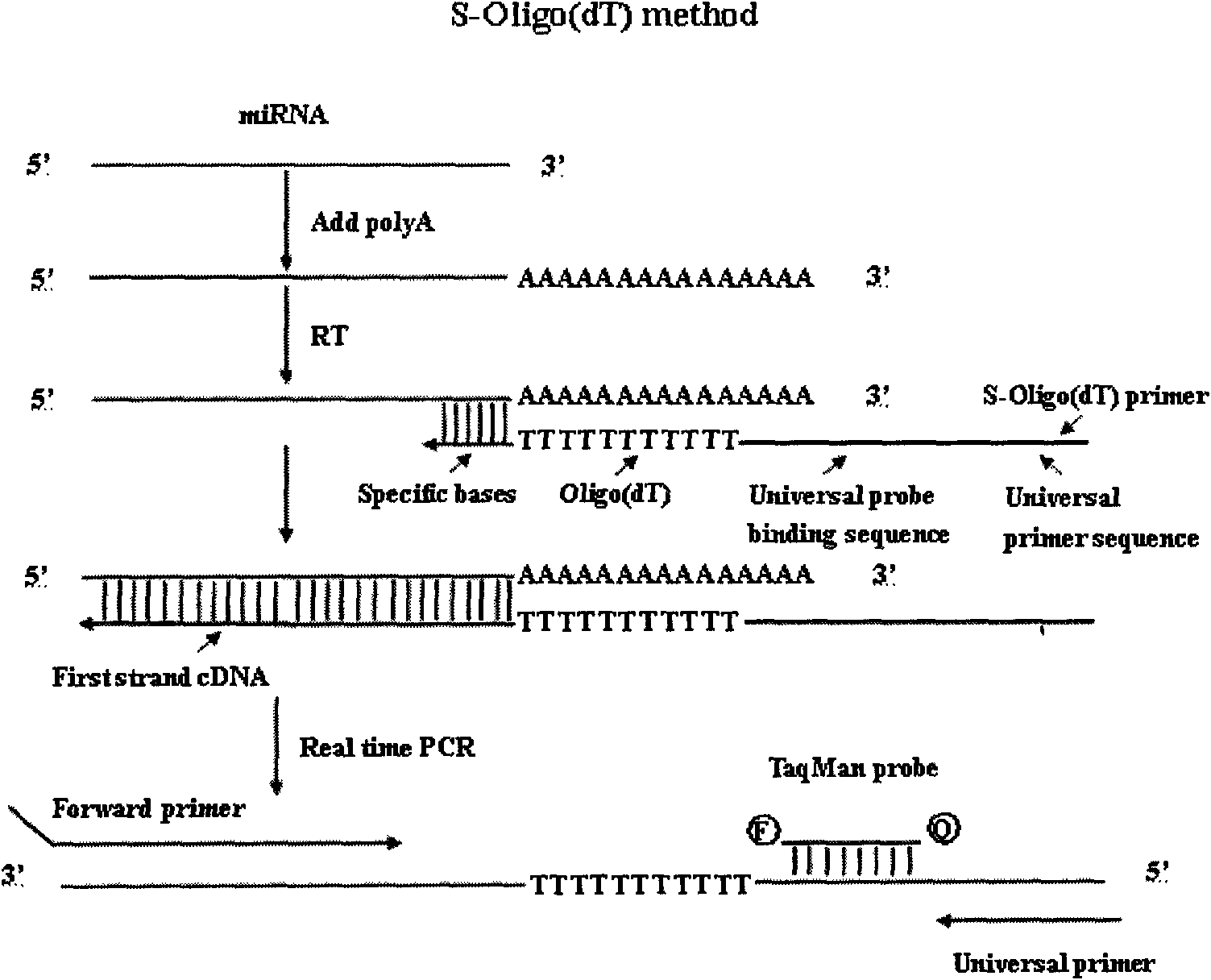
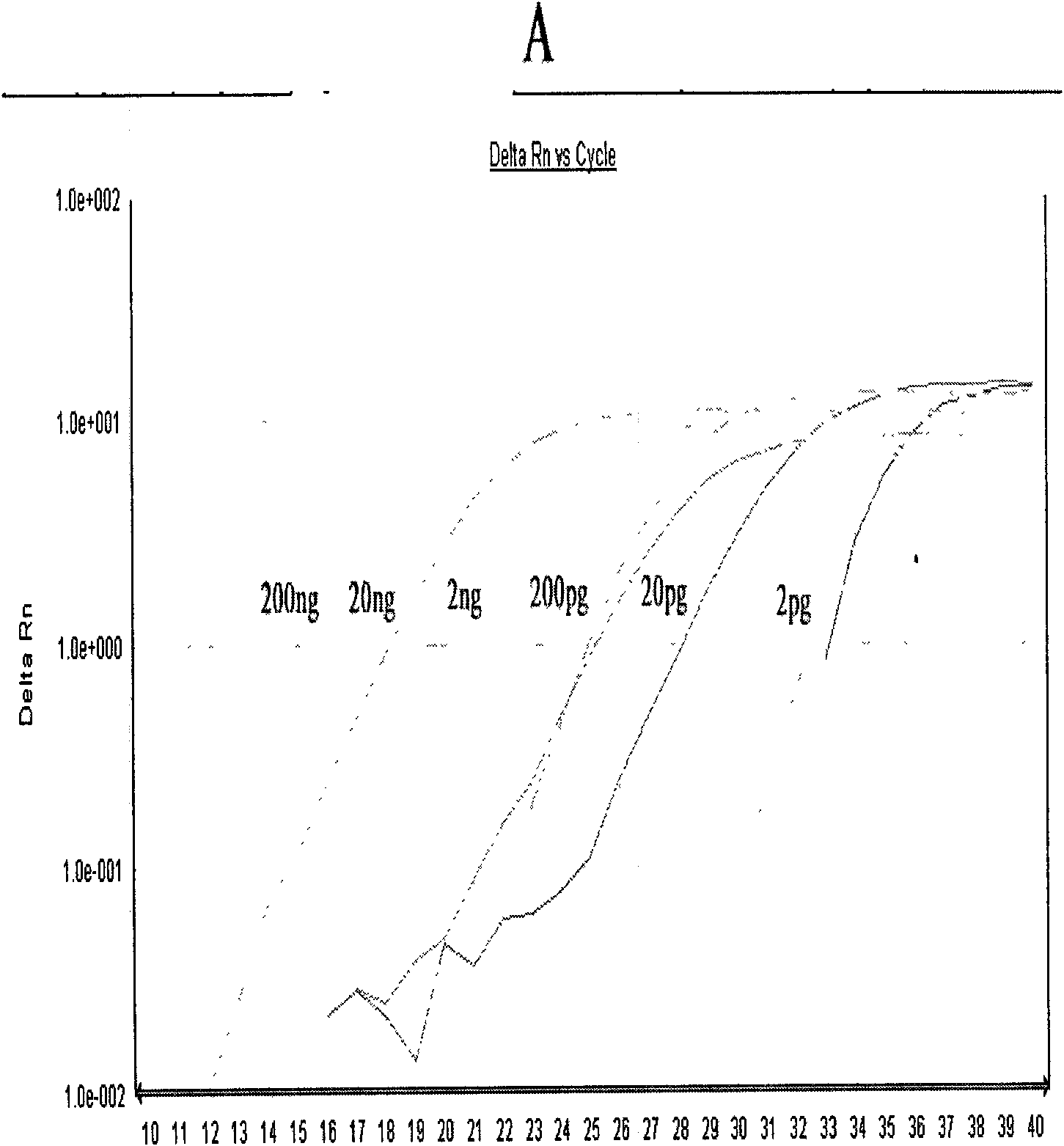
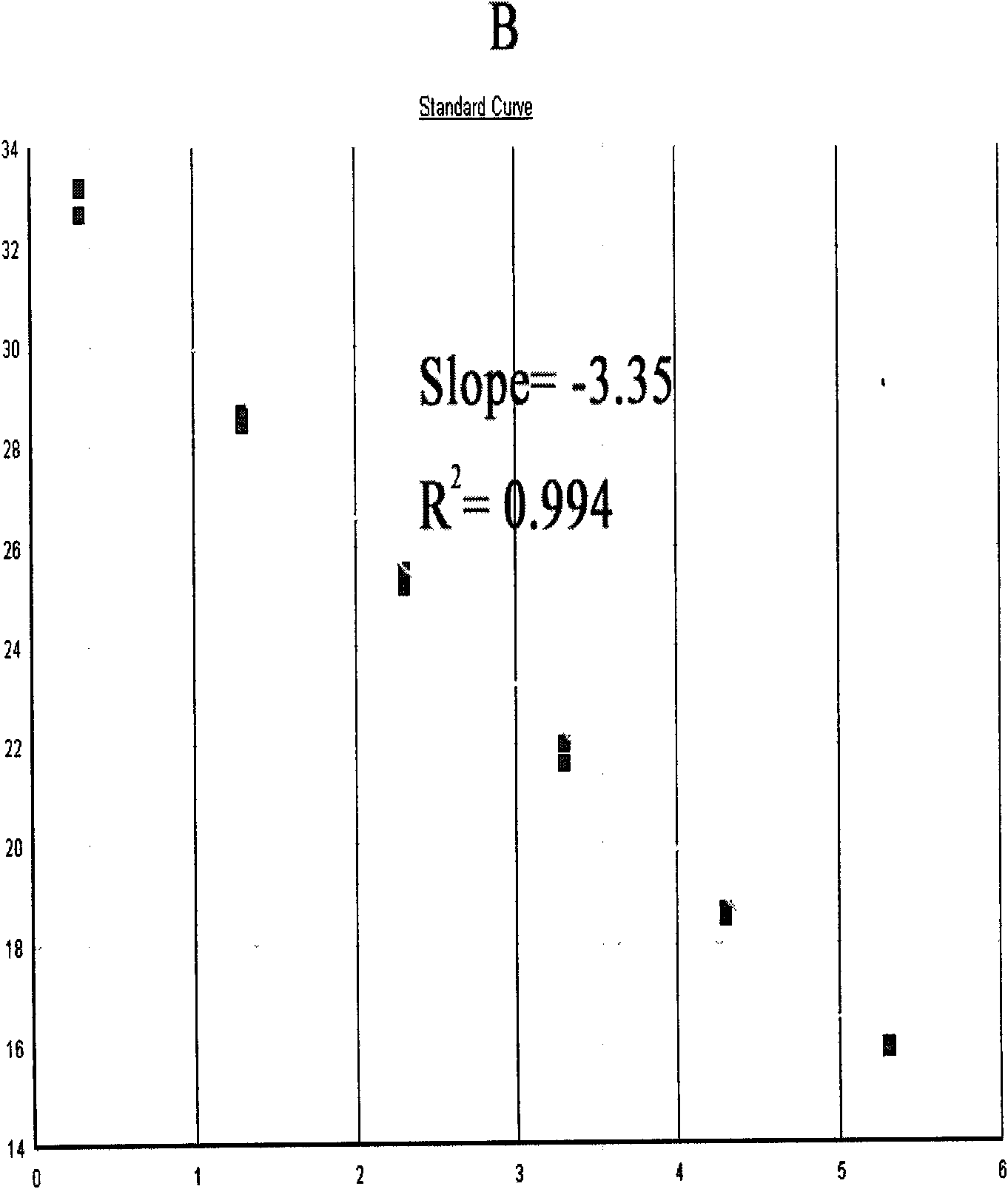
Method and primers for detecting mi ribonucleic acid (miRNA) and application of method
A quantitative detection, universal primer technology, applied in the field of biomedicine, can solve problems such as expensive, achieve the effect of low cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Quantitative detection of hsa-miR-21miRNA in female cervical cancer Hela cells
[0056] (1) Materials
[0057] 1. Materials:
[0058] Female cervical cancer Hela cells cultured in vitro.
[0059] 2. Reagents:
[0060] RNAiso plus (TaKaRa), PolyA Polymerase (EPICENTRE), PrimeScript RTreagent Kit (TaKaRa), SYBR Premix Ex Taq (Peffect Real Time) (TaKaRa), Premix Ex Taq (Perfect Real Time) (TaKaRa).
[0061] 3. Primers and probes:
[0062]
[0063] 4. Instrument:
[0064] GeneQuant pro RNA / DNA quantitative analyzer, ABI PCR instrument (2720), ABIreal-time PCR instrument (PRISM 7300).
[0065] (2) Method
[0066] 1. RNA extraction
[0067] RNA was extracted from Hela cells cultured in vitro with RNAiso plus reagent. The OD260 / 280 and RNA concentration of the extracted Hela cell total RNA were measured by a spectrophotometer, and the integrity of the RNA was analyzed by agarose gel electrophoresis.
[0068] 2. RNA tailing reaction
[0069] The total RNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com