Method and device for assembling genome sequence
A technology for assembling genomes and reference genomes, which is applied in the field of bioinformatics, can solve problems such as bottlenecks and short fragment sequences, and achieve the effects of improving effects, increasing length, and improving assembly efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
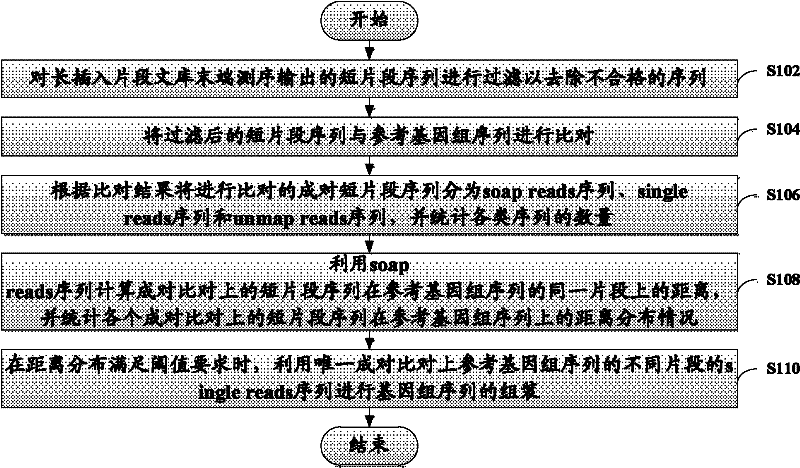
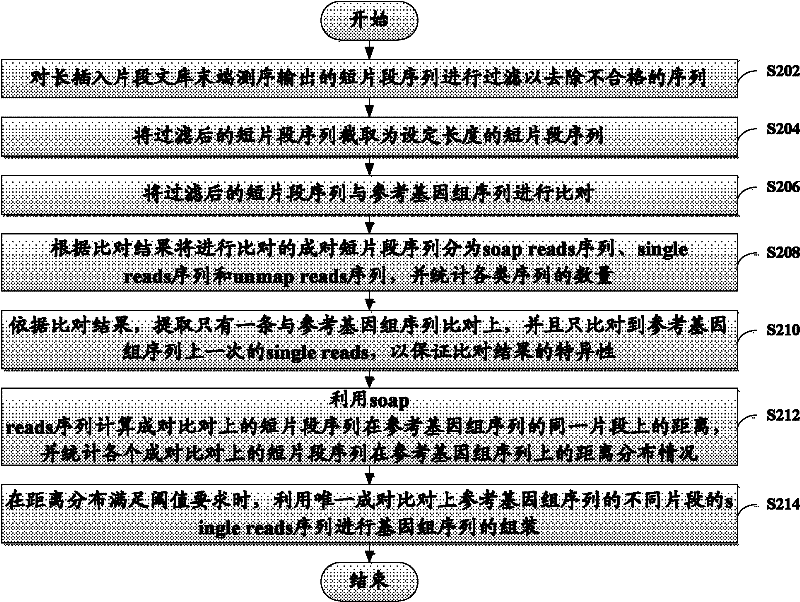
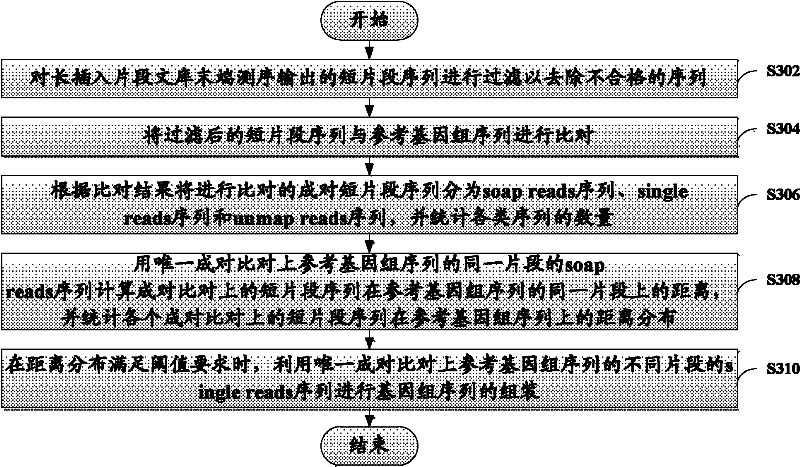
[0027] The present invention will be described more fully hereinafter with reference to the accompanying drawings, in which exemplary embodiments of the invention are illustrated. The exemplary embodiments of the present invention and their descriptions are used to explain the present invention, but do not constitute an improper limitation of the present invention.
[0028] The following description of at least one exemplary embodiment is merely illustrative in nature and in no way taken as limiting the invention, its application or uses.
[0029] Fosmid and Bacterial Artificial Chromosome (BAC) are large-segment clones available in genome research. BAC can usually insert fragments of about 100kb-200kb, and fosmids can generally insert fragments of about 40kb. BAC and fosmid not only have insert lengths characteristics, but also have very good stability, so they are important tools for genomics research, and play an important role in gene map cloning, gene analysis, structural...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com