Method for promoting gene cloning efficiency
A gene and efficiency technology, applied in DNA preparation, recombinant DNA technology, etc., can solve the problems of complex steps, low efficiency in finding main effect factors and optimal reaction conditions, time-consuming and labor-intensive, etc., and achieve the effect of improving the success rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
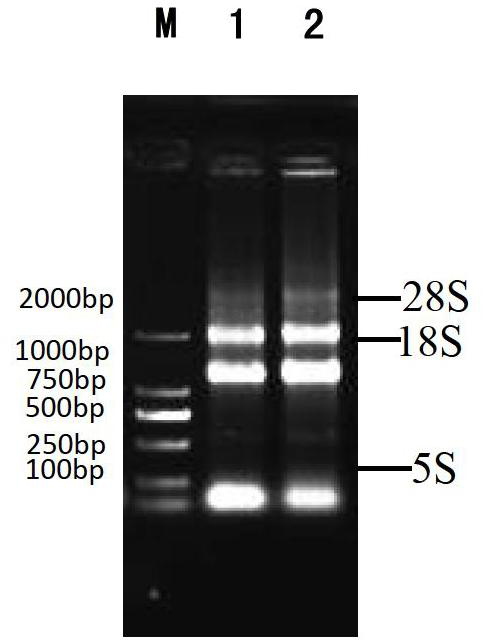
[0028] 1) Extract the total RNA from the liver and head kidney of healthy red-spotted grouper, and test its quality by 1% agarose gel electrophoresis. The buffer system is 0.5×TBE. The results are as follows figure 1 As shown, the 5S, 18S and 28S bands are obvious, indicating that the quality of the extracted RNA is good.
[0029] 2) The first-strand cDNA was synthesized by reverse transcription according to the instructions of the PrimeScriptII 1st Strand cDNA Synthesis kit from TaKaRa.
[0030] 3) Use primers F43 and R167 to detect the product of step 2. The 25 μL system composition of the PCR reaction is: 10×Buffer (containing Mg 2+ ) 2.5 μL, dNTP (dATP, dTTP, dGTP, dCTP each 2.5 mM) 0.5 μL, primers F47 (10 μM) and R167 (10 μM) each 0.5 μL, Taq enzyme (2.5 U / μL) 0.5 μL, template (step 3 The product was diluted 100 times) to 5 μL, and made up to 25 μL with double distilled water; the reaction program was: 95°C for 5 min→(94°C for 30 s, 52°C for 30 s, 72°C for 30 s)×30→72°C ...
Embodiment 2
[0033] 1) Use RNase to degrade the RNA in the reverse transcription product derived from liver tissue.
[0034] 2) Purify cDNA by alcohol precipitation.
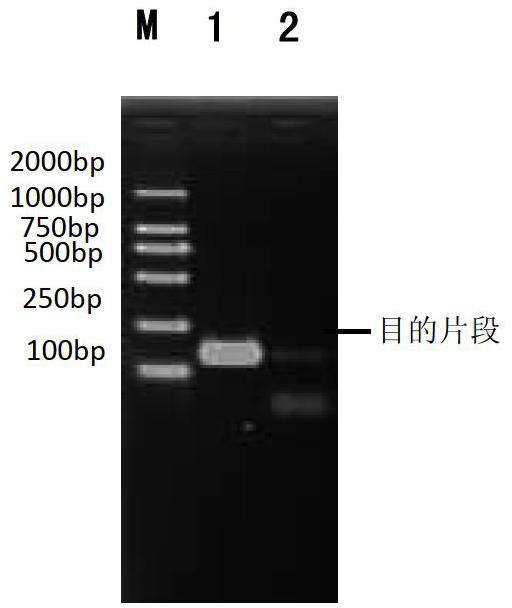
[0035] 3) Use primers F43 and R167 to detect the purified product in step 2. The reaction procedure and system composition are the same as step 4 in Example 1. Test results such as image 3As shown, the results show that there is a brighter positive band, which proves that the purification step has been successfully completed.
Embodiment 3
[0037] 1) Using the first-strand cDNA of red-spotted grouper liver as a template, TdT terminal transferase and dATP were used for homopolymer tailing.
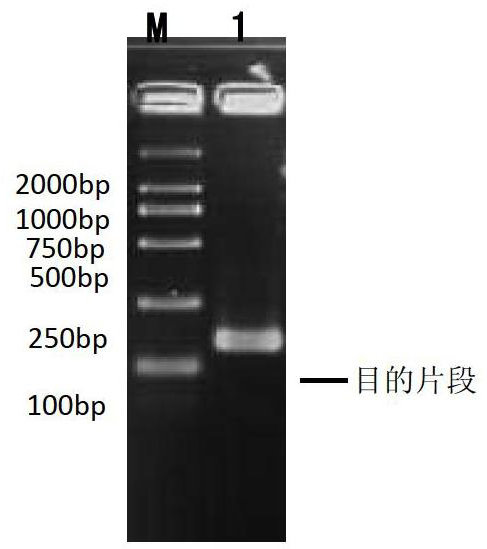
[0038] 2) Choose five annealing temperatures of 50°C, 52°C, 54°C, 56°C, and 58°C for the first round of RACE 5' PCR amplification, and the amplification primers are specific primers R167 (5'-CTGAGCCGTCCTGGCACGCAATG-3'), Linker primer RA (5'-AAGCAGTGGTATCAACGCAGAGT-3') and linker primer oligo-dT-RA with oligo-dT, the sequence is 5'-AAGCAGTGGTATCAACGCAGAGTAC(T) 30 VN-3'. After the product was detected by using primers F43 and R167 with the reaction conditions of step 4 of Example 1, it was analyzed by 1.5% agarose gel electrophoresis, and the results were as follows: Figure 4 As shown, the detection bands of amplified products at 50°C and 58°C are the weakest, while the brightness of other temperature gradients are relatively strong, indicating that temperature is one of the important factors affecting the reaction results. I...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com