PCR (polymerase chain reaction) method for quickly and efficiently detecting DNA (deoxyribonucleic acid) methylation modification after sulfite treatment
A sulfite and methylation technology, applied in the field of molecular biology, can solve problems such as inability to amplify, difficult PCR amplification, cumbersome and complicated, etc., and achieve the goal of expanding the combination range, improving amplification efficiency, and high amplification efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
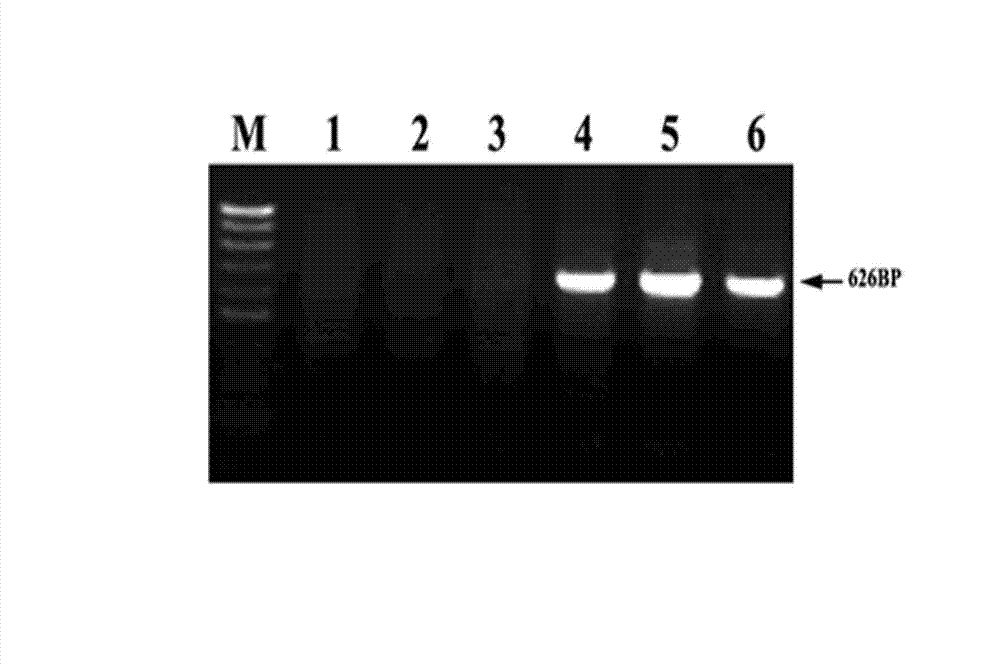
[0045] Example 1 PCR method for detecting the DNA methylation modification of the left LTR of the retrotransposon Tos17 located on chromosome 10 in the rice genome after sulfite treatment
[0046] a) Design internal and external primers
[0047] 1) According to the clone number AC087545 of Tos17 on Genebank, download the Tos17 sequence on the NCBI website, and the left LTR sequence is 138bp, and then download the 600bp sequences of the upstream and downstream of the left LTR sequence through the NCBI website, and use the primer design software pimer5 Primers were designed according to the following method;
[0048] 2) The primer size is 28-35nt, and A and T should be avoided for more than 3 consecutive repetitions;
[0049] 3) The end of the forward primer cannot be C;
[0050] 4) The forward primer is less than 5 C, and the reverse primer is less than 5 G;
[0051] 5) C in the forward primer replaces C / T with the merged base Y; G in the reverse primer replaces G / A with t...
Embodiment 2
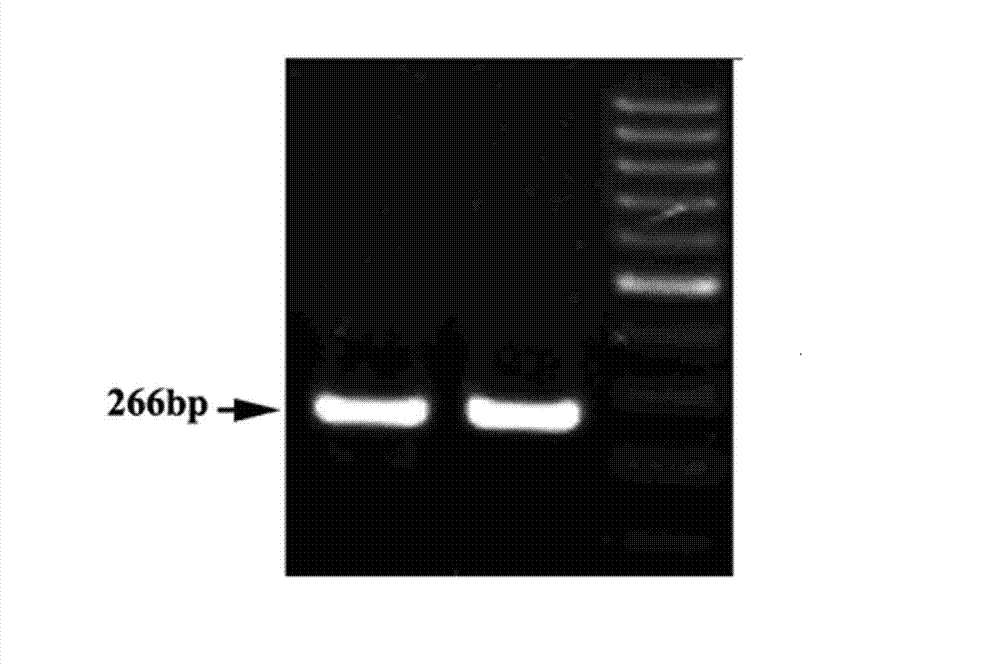
[0071] Example 2 PCR method for detecting the DNA methylation modification of the left LTR of the retrotransposon Tos17 located on chromosome 7 in the rice genome after sulfite treatment
[0072] a) Design internal and external primers
[0073] 1) According to its clone number AP005292 on Genebank, this sequence was downloaded from the NCBI website, which is located at the position of 186486-190599 on chromosome 7, and then the sequence of 600 bp upstream and downstream of the left LTR sequence was downloaded through the NCBI website, which is located at At 185886-187086 of chromosome 7, use the primer design software pimer5 to design primers according to the following method;
[0074] 2) The primer size is 28-35nt, and A and T should be avoided for more than 3 consecutive repetitions;
[0075] 3) The end of the forward primer cannot be C;
[0076] 4) The forward primer is less than 5 C, and the reverse primer is less than 5 G;
[0077] 5) C in the forward primer replaces...
Embodiment 3
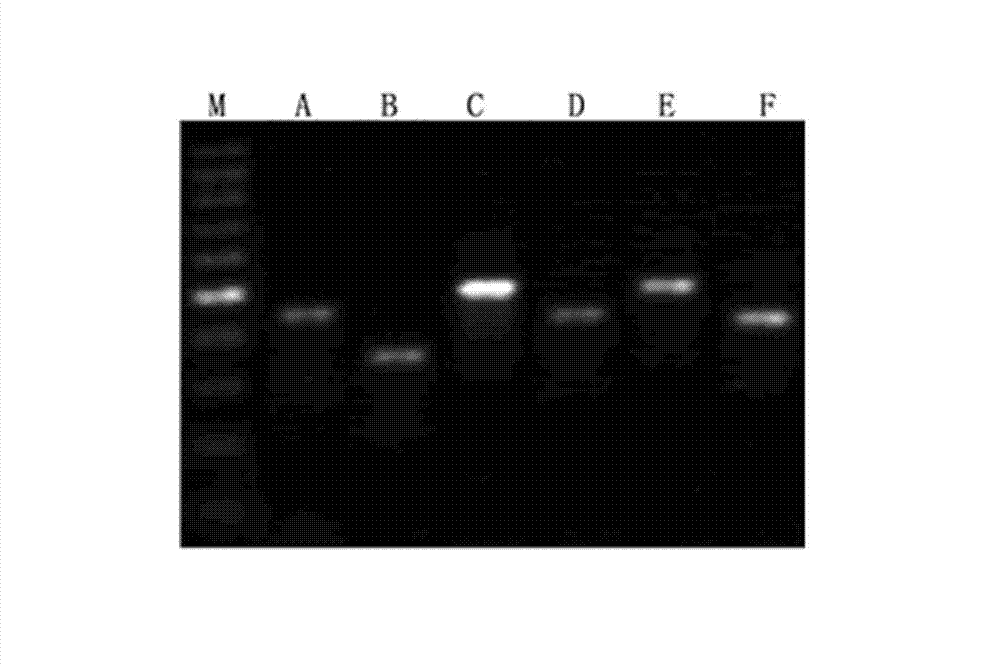
[0097] Example 3 PCR method for detecting DNA methylation modification of sequence AP001551 in GENE BANK after sulfite treatment
[0098] a) Design internal and external primers
[0099] 1) According to its clone number AP001551 on Genebank, download this sequence on the NCBI website, which is located at the position 59722-64876 of chromosome 1, and then download the sequence of 300 bp upstream and downstream of the left sequence through the NCBI website, which is located at a At 59422-60022 of chromosome number, use the primer design software pimer5 to design primers according to the following method:
[0100] 2) The primer size is 28-35nt, and A and T should be avoided for more than 3 consecutive repetitions;
[0101] 3) The end of the forward primer cannot be C;
[0102] 4) The forward primer is less than 5 C, and the reverse primer is less than 5 G;
[0103] 5) Using the merger method, the C in the forward primer is replaced by the merged base Y for C / T; the G in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com