Method for analysis of cellular DNA content
一种细胞、含量的技术,应用在测定生物细胞的核酸量领域,能够解决粘附细胞脱落、损害单个细胞DNA含量的量化、细胞损失等问题,达到可靠结果的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
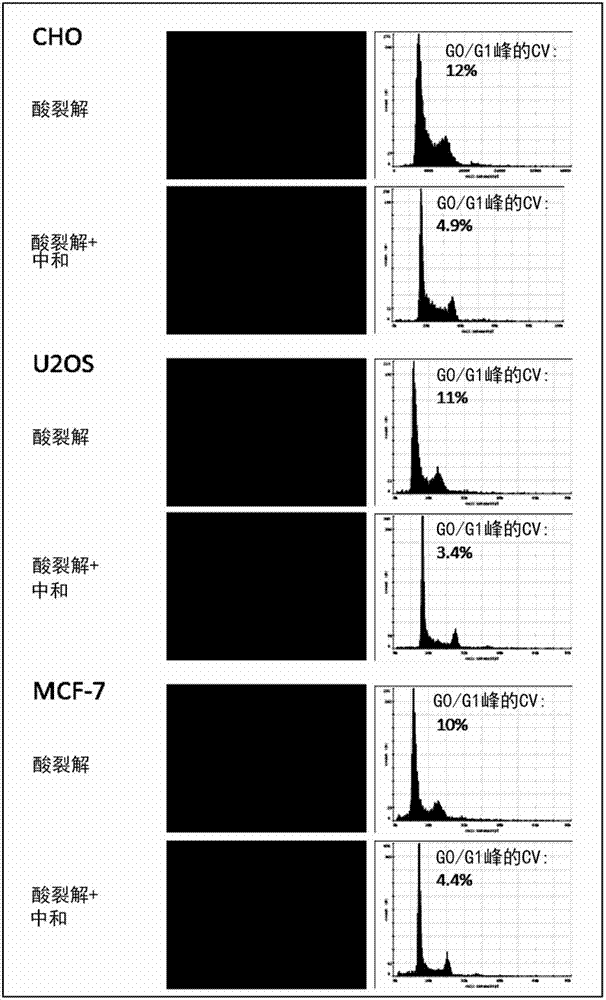
[0312] Example 1. Nuclei Released by Acid Cleavage and Used for Accurate and Precise DNA Quantification
[0313] The DNA content of three adherent cell lines CHO, MCF-7 and U2OS was measured by staining nuclei released by lysis with DAPI (4',6-diamidino-2-phenylindole) acid.
[0314] Materials and methods:
[0315] Adherent cell lines were grown to 80-90% confluency in RPMI+10% FCS in T25 flasks. To release nuclei directly from attached cells, wash the T25 flask once with 5 mL of PBS and then at 37 °C in 0.65 ml of lysis buffer (50 mM citric acid (pH 2.20), 1% Triton X-100, 1 μg / ml DAPI) Incubate in presence for 5 minutes. Fluorescence of released nuclei was either analyzed directly by quantitative imaging cytometry or neutralized by adding an equal volume (0.65 ml) of stabilization buffer (250 mM citrate (pH 8.3), 1% Triton X-100) prior to imaging. and. Quantitative imaging cytometry was performed using a Nucleocounter NC-3000 (Chemometec).
[0316] result:
[0317]...
Embodiment 2
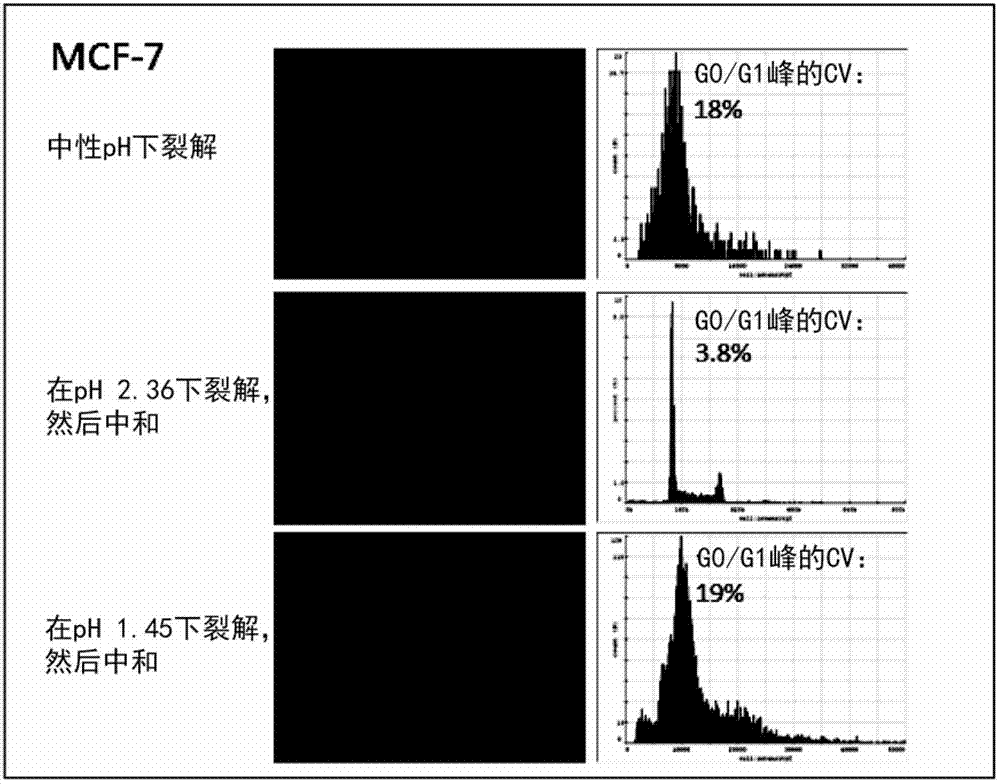
[0321] Example 2, Determination of the Optimal pH of Nuclear Release, Deaggregation and Uniform DNA Staining
[0322] Three adherent cell lines CHO, MCF-7 and U2OS were used to determine the optimum pH for the following three parameters:
[0323] Efficient release of nuclei (>90 nuclei released from attached cells)
[0324] Little or no aggregation of released nuclei
[0325] Uniform DNA staining that provides high accuracy and precision in DNA content measurement
[0326] Materials and methods:
[0327] Adherent cell lines were grown to 80-90% confluency in RPMI+10% FCS in 6-well plates (Nunclon surface, Nunc). To release nuclei directly from attached cells, wash each well once with 1 mL of PBS and dissolve in 0.25 ml of lysis buffer (0.5% Triton X-100, 1 μg / ml DAPI dissolved in 200 mM citric acid (pH 2.00) at 37°C. , 100 mM citrate buffer (pH 2.22-6.80) or dissolved in 400 mM phosphoric acid (pH 1.45)) for 5 minutes. The released nuclei were neutralized by the additio...
Embodiment 3
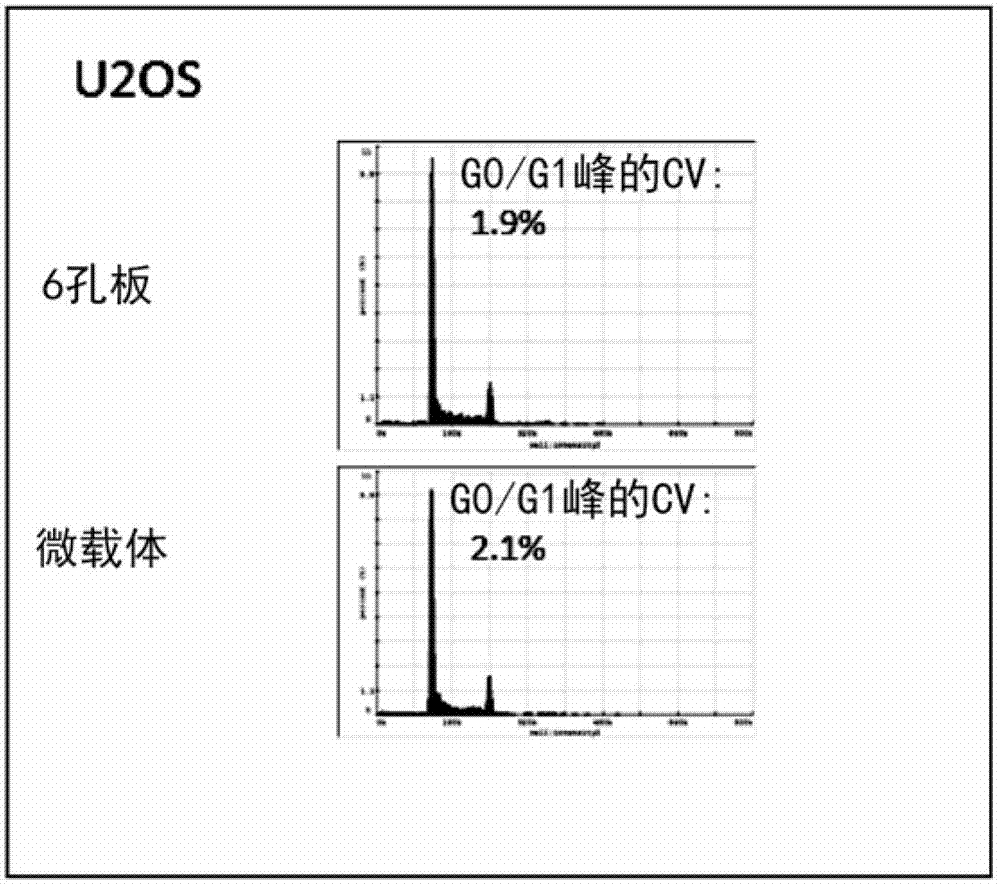
[0339] The influence of embodiment 3, DAPI concentration and dyeing method
[0340] Whether the accuracy and precision of DNA staining using the adherent cell line U2OS could be further improved by performing nuclear staining in the neutralization step and not in the lysis step.
[0341] Materials and methods:
[0342] U2OS cells were grown to 80-90% confluency in RPMI+10% FCS in 6-well plates. To release nuclei directly from attached cells, wash the wells once with 1 mL of PBS and then in 0.25 ml of lysis buffer (100 mM citrate buffer (pH 2.25), 37 °C, in the presence or absence of DAPI, 0.5% Triton X-100) for 5 minutes. The released nuclei were neutralized by incubation with 0.25 ml stabilization buffer (500 mM citrate, pH 8.3) at 37°C in the presence or absence of DAPI. See Table 2 for details on the experimental conditions, which shows the effect of DAPI concentration and staining method. Fluorescence of nuclei was analyzed by imaging cytometry using a Nucleocounter ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com