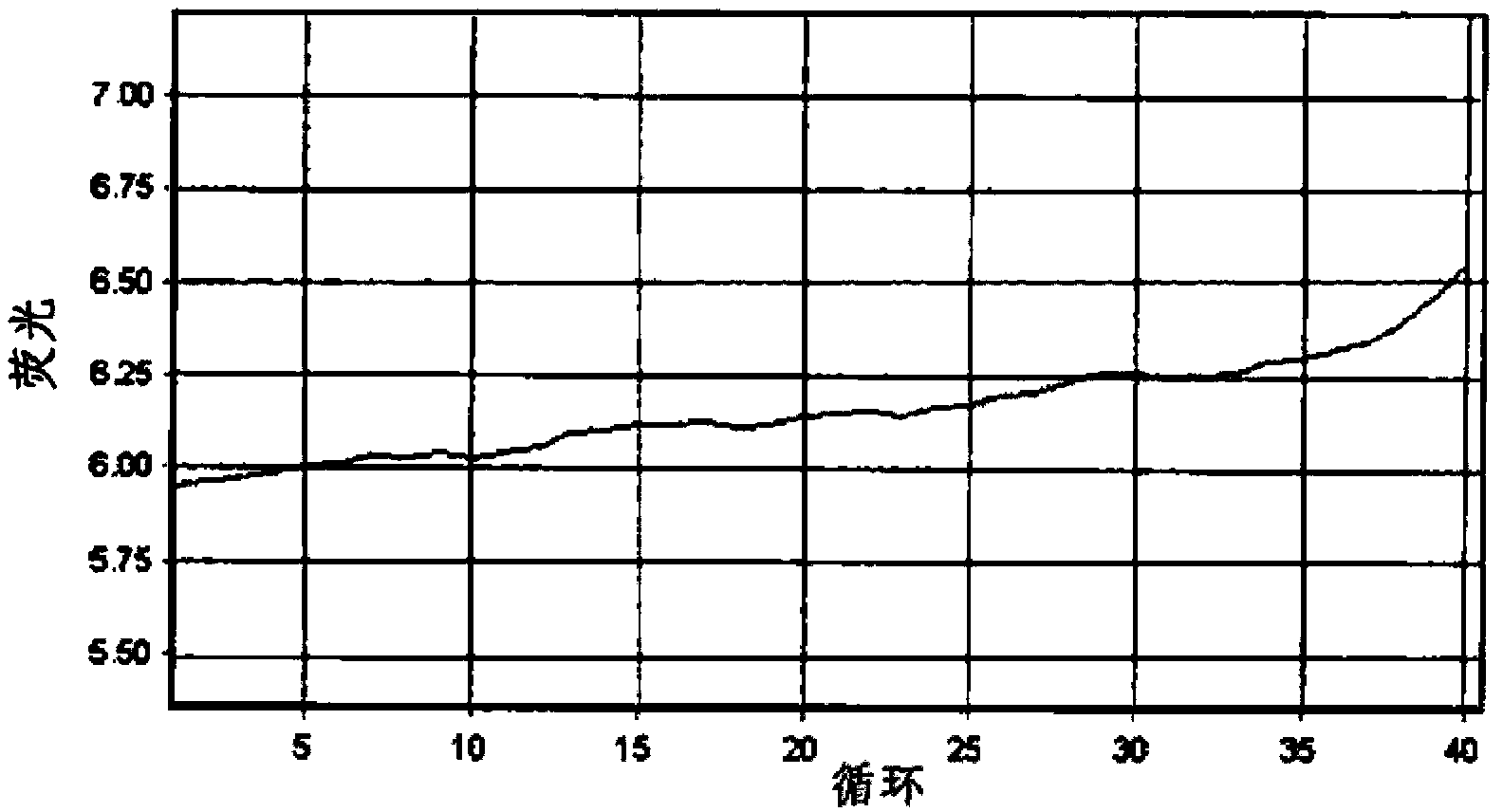
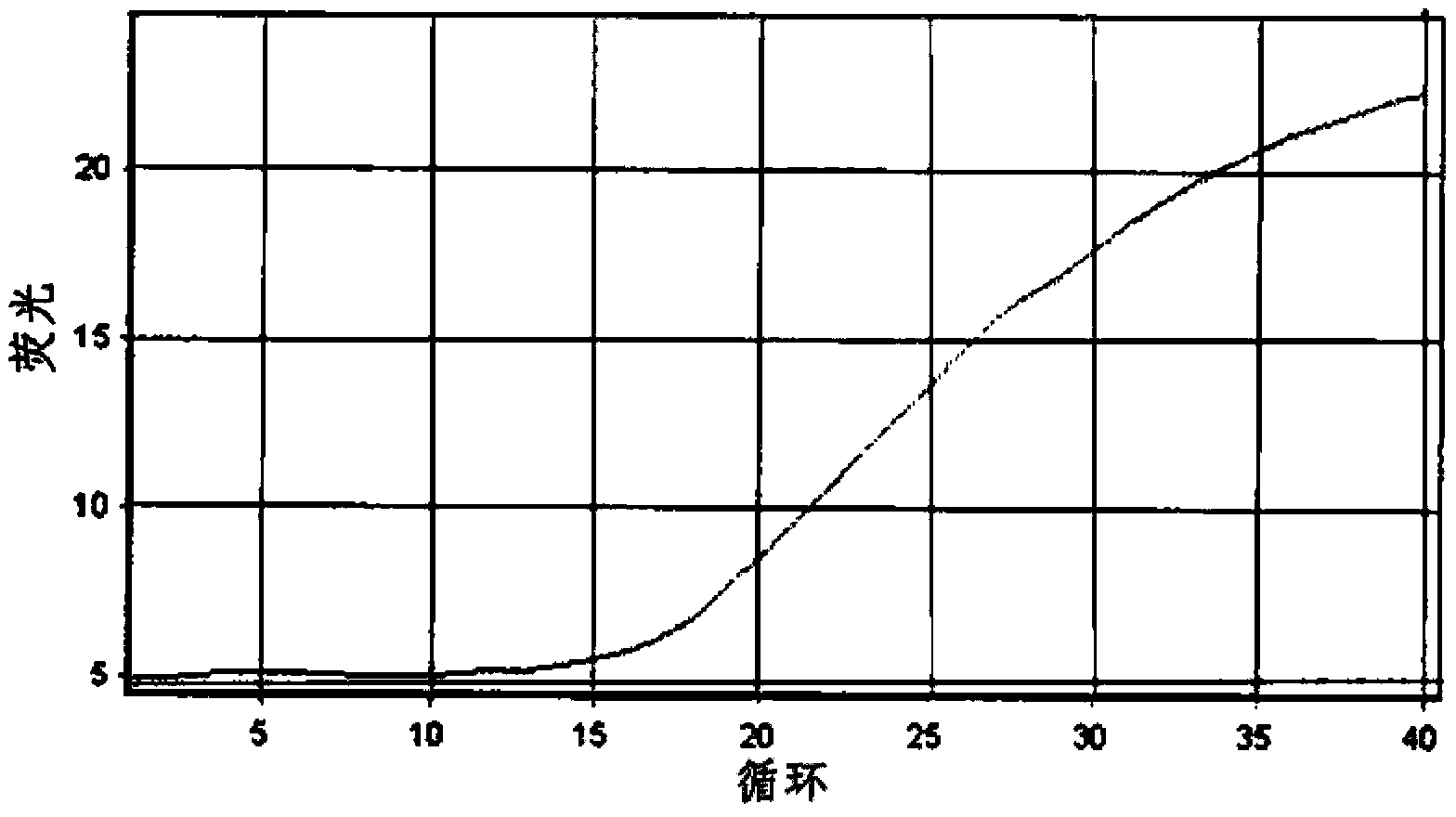
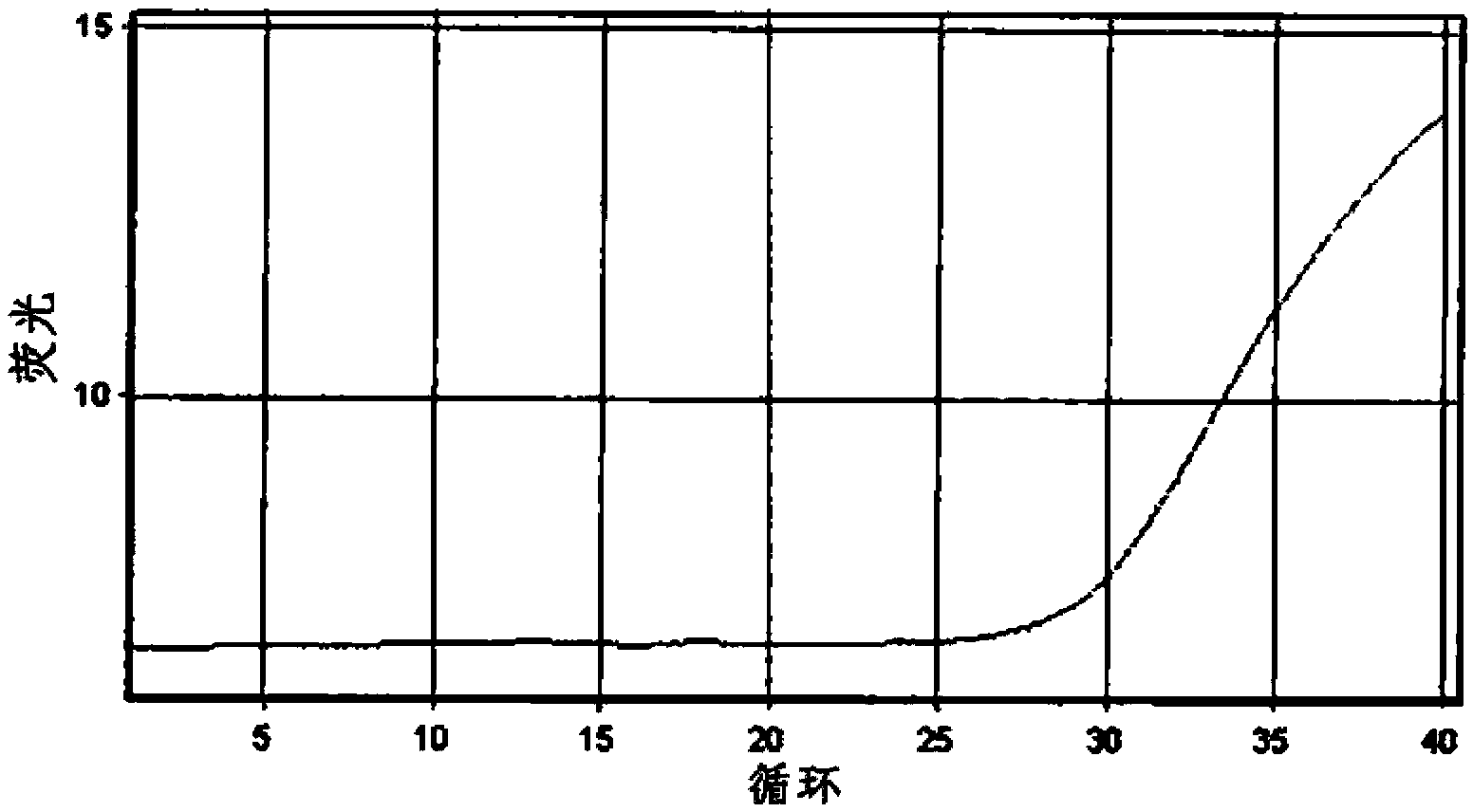
Method for the detection of Mycobacterium tuberculosis and nontuberculous mycobacteria by the use of dual real-time polymerase chain reaction
A technology of mycobacterium tuberculosis and mycobacteria, applied in the direction of microorganism-based methods, biochemical equipment and methods, biological testing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0169] Example 1: Isolation and Detection of Mycobacterium Tuberculosis Complex and Nontuberculous Mycobacterium 1
[0170] 1. Detection target and primer design
[0171] The target genes to be detected are the IS6110 gene of Mycobacterium tuberculosis complex (MTC: Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium africanum, and Mycobacterium microti) and the 16S rRNA of nontuberculous mycobacteria (NTM) Gene. Primers and Taqman probes designed using the Primer3 program were used in the detection of the target gene.
[0172] (1)MTC
[0173] 1) Target gene: IS6110
[0174] 2) Primers
[0175] a. Forward primer: 5'-cgaactcaaggagcacatca-3' (SEQ ID NO: 1)
[0176] b. Reverse primer: 5'-agtttggtcatcagccgttc-3' (SEQ ID NO: 2)
[0177] 3) Taqman probe
[0178] 5'-VIC-agtgtggctaaccctgaa-MGB-3' (SEQ ID NO: 3)
[0179] 4) PCR product size: 135bp
[0180] (2)NTM
[0181] 1) Target gene: 16S rRNA
[0182] 2) Primers
[0183] a. Forward primer: 5'-ggyrayctgccctgca...
Embodiment 1-1
[0190]
[0191] (1) DNA separation
[0192]DNA was isolated from 186 mycobacterial species and 78 nontuberculous mycobacterial species (all identified in clinical subjects), as well as 7 standard ATCC mycobacterial species (including Mycobacterium tuberculosis (ATCC25177), Mycobacterium intracellulare (ATCC13950), Mycobacterium scrofula (ATCC19981), Mycobacterium kansasii (ATCC12478), Mycobacterium fortuitum (ATCC6841), Mycobacterium abscessus (ATCC19977) and Mycobacterium avium (ATCC25291)) .
[0193] DNA was isolated from 186 mycobacterial species and 78 nontuberculous mycobacterial species (all identified in clinical subjects), as well as 7 standard ATCC mycobacterial species (including Mycobacterium tuberculosis (ATCC25177), Mycobacterium intracellulare (ATCC13950), Mycobacterium scrofula (ATCC19981), Mycobacterium kansasii (ATCC12478), Mycobacterium fortuitum (ATCC6841), Mycobacterium abscessus (ATCC19977) and Mycobacterium avium (ATCC25291)) .
[0194] Species ident...
Embodiment 1-2
[0203]
[0204] Duplex real-time PCR was performed in the same manner as in Example 1-1, except that the nucleotide sequence of SEQ ID NO: 6 was used as the reverse primer NTM-1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com