A gene chip for detection of individualized treatment of Helicobacter pylori infection and its application
A technology for detecting gene chips and Helicobacter pylori, which is applied in the field of medical inspection and gene chips, individualized treatment of Helicobacter pylori infection and detection of gene chips
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: Preparation of a gene chip for detection of individualized treatment of Helicobacter pylori infection
[0046] 1. Preparation of specific primers
[0047] Select the clarithromycin-resistant mutation sites A2142G and A2143G in the Helicobacter pylori 23s gene; the quinolone-resistant catastrophe sites Ash87 and Asp91 in the Helicobacter pylori gyrA gene; 2C19*2(G6981A) and 2C19*3(G636A) as detection targets
[0048] Specific primers were designed according to the standard (see Table 1). All downstream primers were 5'-end labeled with biotin.
[0049] Table 1 Specific primers and their corresponding amplification target gene types
[0050]
[0051] 2. Design and screen probes for human proton pump inhibitor metabolism-related sites and Helicobacter pylori drug-resistant sites
[0052] Wild-type and mutant SNP typing probes were designed for A2142G, A2143G, N87K, D91G / Y / N, G681A, and G636A, respectively (as shown in Table 2). Simultaneously design matri...
Embodiment 2
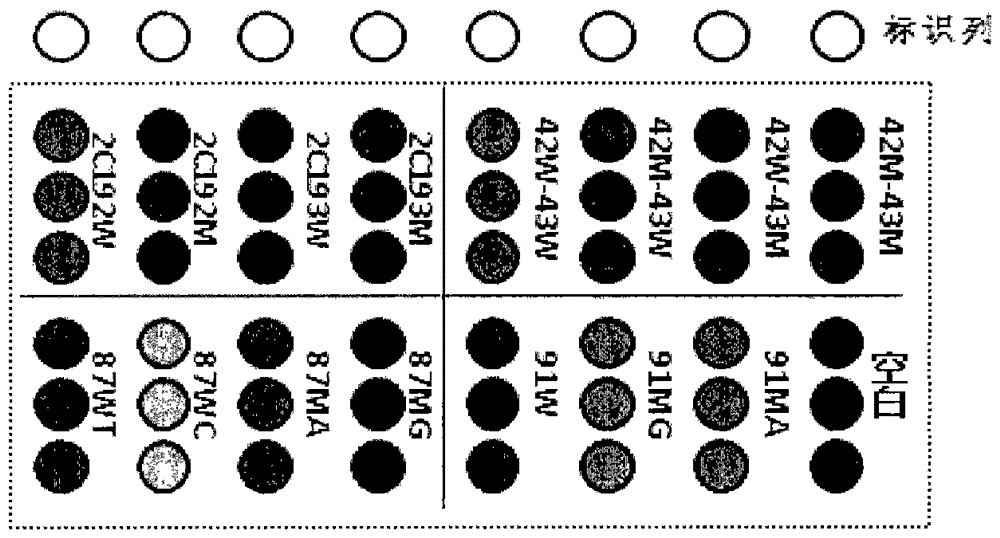
[0071] Example 2: Determination of Gene Chip Positive Judgment Criteria
[0072] All result judgments are based on the signal value calculated by the software, which reflects the gray value of the probe. The chip matrix control points, negative quality control points, and the positive value interpretation standards of the human genome and Helicobacter pylori drug resistance site probes are all calculated according to statistical methods. For the two probes of CYP450-2C19*2 (2C19*2-W and 2C19*2M), when the average gray value of a certain probe is twice or more than that of the other, it is judged that the probe is positive ; When the ratio of the average gray value of the two is less than 2, it is judged that the locus is heterozygous. The interpretation criteria of the two probes (2C19*3-W and 2C19*3M) of CYP450-2C19*3 are the same as those of CYP450-2C192. For the 4 probes (42W-43W, 42M-43W, 42W-43M and 42M-43M) of the clarithromycin-resistant site, when the average gray va...
Embodiment 3
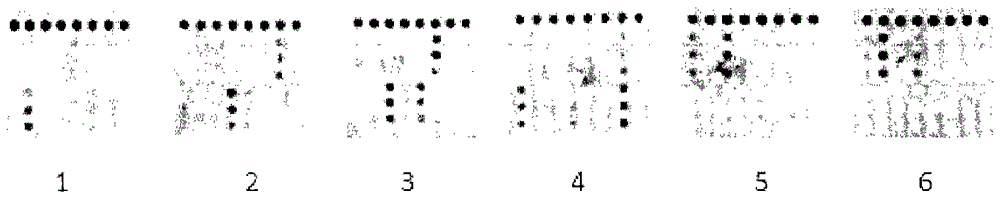
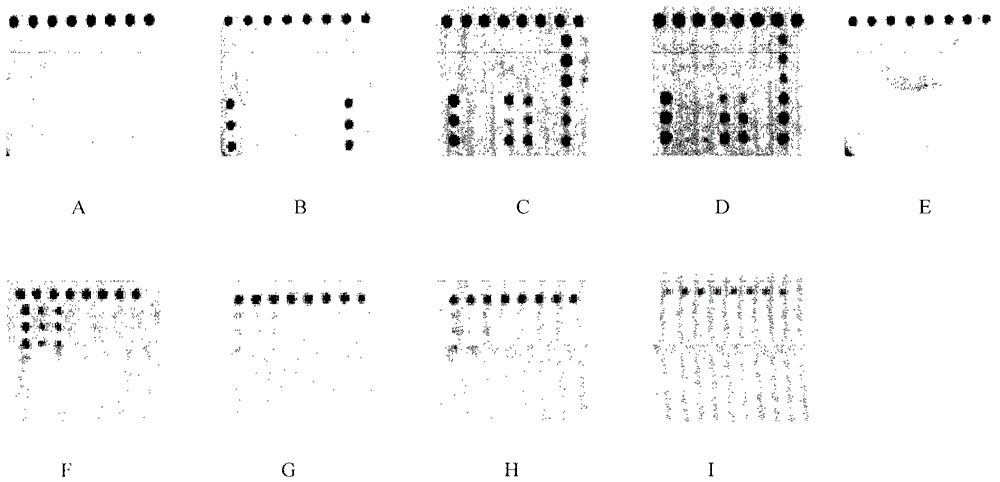
[0073] Example 3: Specificity evaluation of gene chip detection for individualized treatment of Helicobacter pylori infection
[0074] Specificity is the most important assessment index of a diagnostic method. The present invention uses optimized systems and conditions to detect various strains and human genomes with common mutations. The chip detection results are shown in the attached figure 2 . It can be seen from the figure that the present invention can correctly distinguish bacterial strains with common mutations and human genome, and the specificity is good.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com