Primer and identifying method for identifying different genetic collateral series of aphelinid
A technology of solar bees and branches, applied in the field of agricultural biology, can solve problems such as high chance of polymorphism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 3
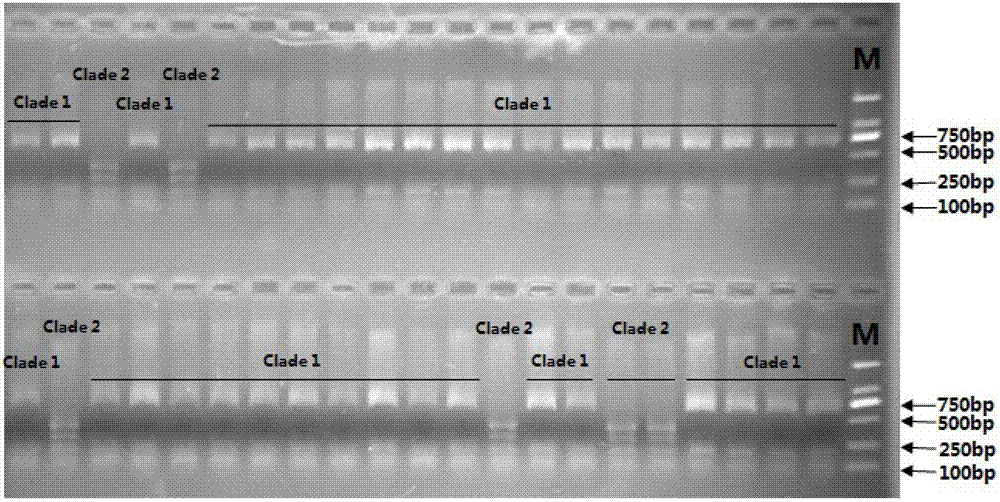
[0035] The Sunbee described in Example 3 was collected in Changzhi City, Shanxi Province in 2007; the above Sunbee was tested, and the Sunbee collected in Changzhi City, Shanxi Province was genetic branch 1 and genetic branch 2.
Embodiment 4
[0036] The Sunbee described in Example 4 was collected in Dalian City, Liaoning Province in 2007; the above Sunbee was tested, and the Sunbee collected in Dalian was evenly divided into genetic lineage 2.
[0037] The Alu I endonuclease, Tris-HCl, ethylenediaminetetraacetic acid, and sodium lauryl sulfate described in the examples were all purchased from Shanghai Bioengineering Company, and other reagents were all commercially available products.
Embodiment 1
[0039] (i) Extraction of Sunbee Genomic DNA
[0040] Place a single sunbee in a 0.2ml centrifuge tube containing 60 μl of alkaline lysate: 50 mmol L -1 Tris-HCl (pH8.0), 20mmol·L -1 NaCl, 1mmol L -1 EDTA (ethylenediaminetetraacetic acid) and 1% SDS (sodium dodecyl sulfate) are fully ground and homogenized with a sealed gun head, placed in a water bath at 65°C for 15 minutes, and then placed in a water bath at 95°C for 10 minutes to obtain Sunbee Genomic DNA Solution.
[0041] (ii) PCR amplification of COI gene of sunbee
[0042] Carry out PCR amplification with the solar bee genomic DNA solution as a template to obtain PCR amplification products;
[0043] The PCR amplification system is:
[0044] Sunbee Genomic DNA Solution: 3μl; 20μM Primer: 0.5μl; 5U / μl Taq Enzyme: 0.25μl; 10×Taq Buffer: 2.5μl; 10mM dNTP: 0.5μl; ddH 2 0 to 25 μl;
[0045] The primer sequences are as follows:
[0046]Sense primer Clade-F: 5'-TCTCATATAATTTGTAATGAAAG-3';
[0047] Antisense primer Clade...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com