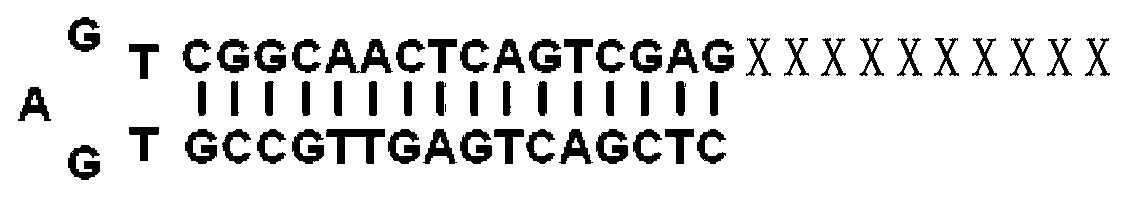Method for obtaining miRNA (Ribose Nucleic Acid) candidate target gene and special reverse transcription primer for method
A technology of reverse transcription primers and candidate targets, applied in DNA/RNA fragments, DNA preparation, recombinant DNA technology, etc., can solve the problems of incomplete genomic data, insufficient throughput, and difficulty in obtaining complete information of sheep skeletal muscle in batches, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1, obtaining the miRNA candidate target gene whose name is mir-133
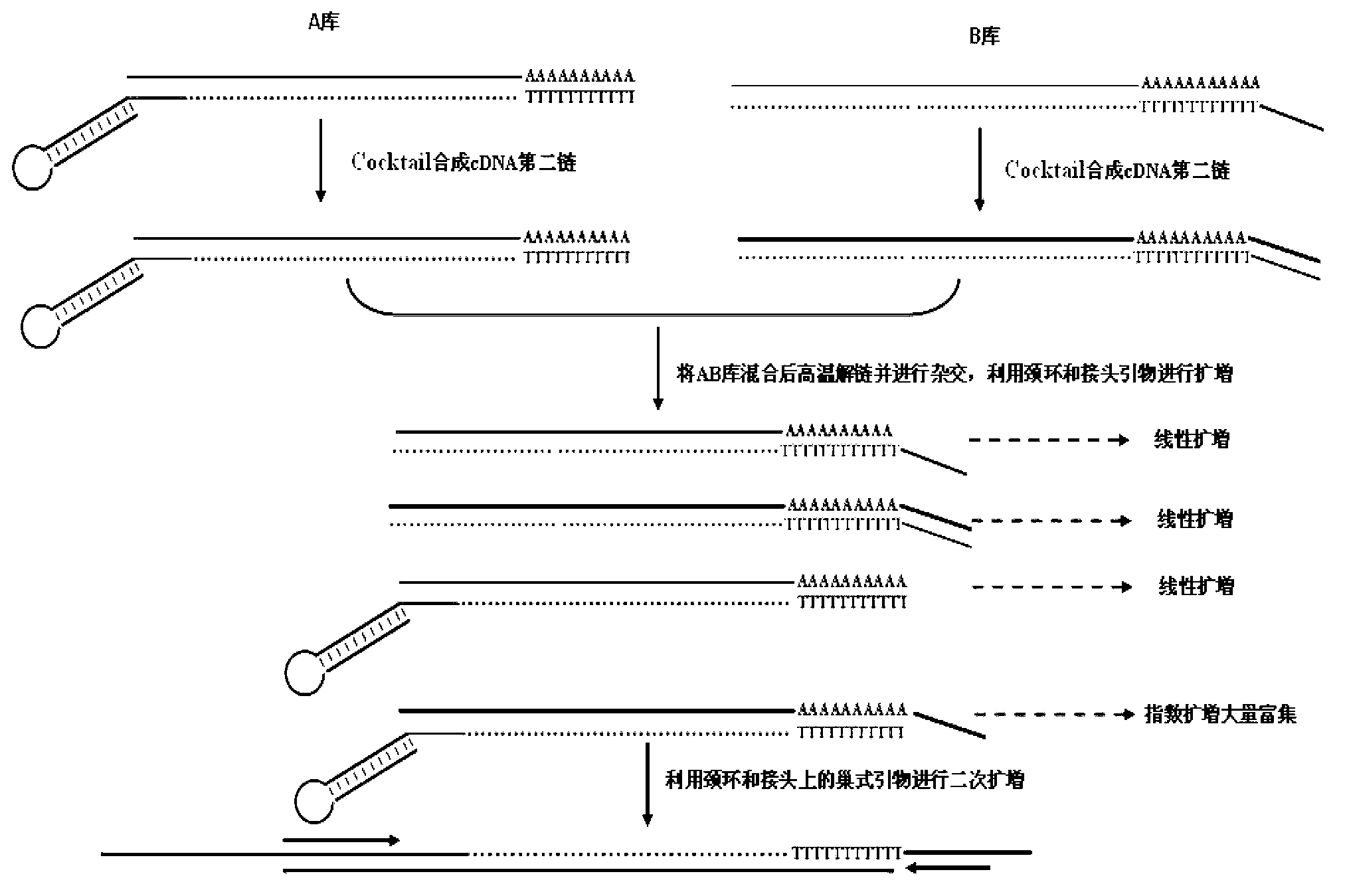
[0050] The miRNA used in this example is mir-133, and its nucleotide sequence in the 3' to 5' direction is GUCGACCAACUU CCCCUGGUUU . The nucleotide sequence of the 5' to 3' direction of the reverse transcription primer A is CTCGACTGAGTTGCCGTGAGTCGGCAACTCAGTCGAG CCCCTGGTTT (The underlined base is the mir-133 seed sequence ) . The nucleotide sequence of reverse transcription primer B is AACGCGTCGCGTCGAGTAGTAGACGTA TTTTTTTTTTTTTTT. The nucleotide sequence of primer A1 is TGAGTCGGCAACTCAGTCGAG , the nucleotide sequence of primer B1 is AGCGCAGCTCATCATCTGCAT . Among them, primer A1 and primer B1 are nested primers.
[0051] The specific method is as follows:
[0052] The total RNA of skeletal muscle of sheep (Chinese Merino sheep) was extracted using commercial Trizol reagent, and the total RNA of skeletal muscle was purified using the Oligotex mRNA purification kit. Divide the purifie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com