Method for synthesizing short single strand deoxyribonucleotide probe
A deoxyribonucleotide, single-stranded technology, which is used in the field of synthesizing short single-stranded deoxyribonucleotide probes, can solve the problem of primer mutations, inability to use single-stranded probes for experiment and diagnosis, and no long single-stranded cleavage. Problems such as forming specific short chains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment one: preparation microRNA probe method one
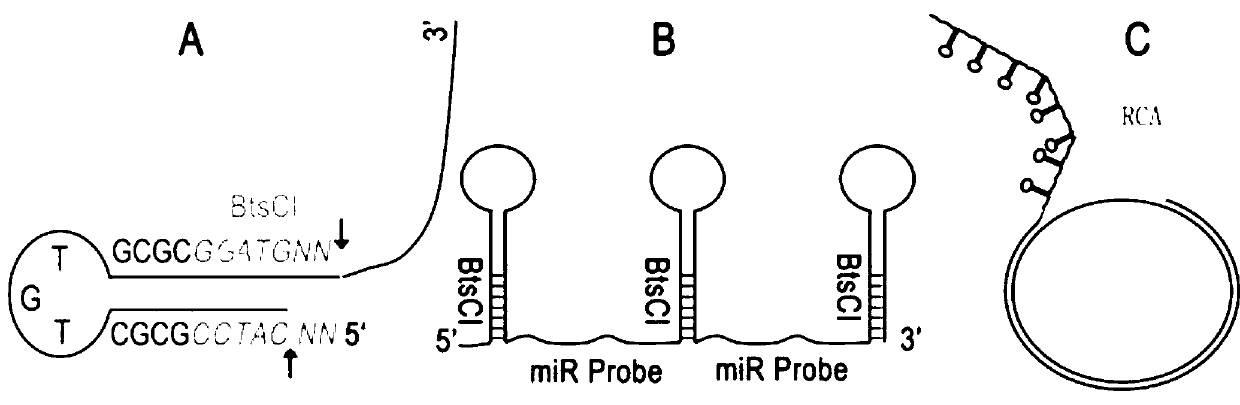
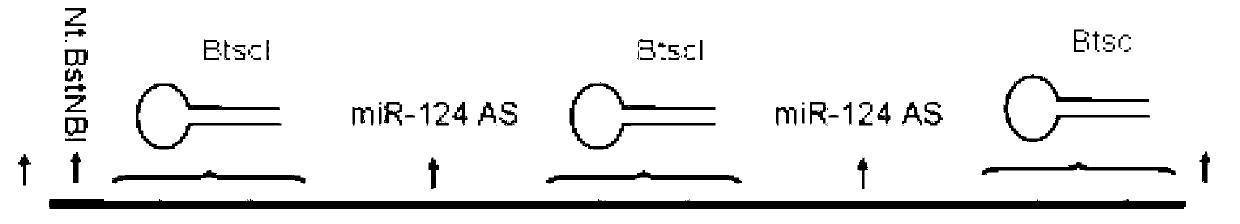
[0042] Step 1: Design the DNA sequence to be amplified. The 5' end is a sequence that can form a neck loop structure containing a type II endonuclease BtsCI site, and the 3' end is an antisense sequence (probe) of a microRNA , or for any sequence of DNA, see figure 1 a. In order to optimize production and follow-up experiments, the above-mentioned neck loop-microRNA probe sequence is superimposed to form a neck loop-probe-neck loop-probe-neck loop sequence, and Nt.BstNBI is added to the 5' end of this sequence The nickase recognition site GAGTGatat, and finally an EcoRI endonuclease recognition site GAATTC is added at both ends, such as figure 1 b.
[0043] The sequence of the mouse microRNA mmu-miR-124-3p (sequence number MIMAT0000134) to be detected is 5'UAAGGCACGCGGUGAAUGCC3' (seq No.3). The sequences designed for the production of miR-124 probes are shown in Table 1.
[0044] Table 1 is used to produce the...
Embodiment 2
[0055] Embodiment two: prepare microRNA probe method two
[0056] Method 2 is roughly the same as method 1, except that in the sixth step of method 1, 0.1mM DIG-dUTP and 0.1mM dTTP are used instead of 0.2mM dTTP, so that the single-stranded DNA synthesized by phi29 DNA polymerase is incorporated into DIG-dUTP . DIG is a marker, which can be digoxin, fluorescein, biotin, isotope, etc. After completing the ninth step of tapping and purifying the DNA, the labeled DNA probes are obtained.
Embodiment 3
[0057] Example 3: Detection of mouse miR-124 with the made target single-stranded DNA probe (i.e. miR-124 probe)
[0058] first step:
[0059] Eight micron thick slices of embryonic day 12.5 mouse brain processed as follows
[0060] 1. Xylene dewaxing twice, 15 minutes each time
[0061] 2. Wash twice with 100% ethanol, 5 minutes each time
[0062] 3. Wash with 70% ethanol for 5 minutes
[0063] 4. Wash with 30% ethanol for 5 minutes
[0064] 5. Wash in deionized water for 5 minutes
[0065] 6. Wash twice in 1x PBS, 5 minutes each
[0066] 7.4% PFA fixed on ice for 20 minutes
[0067] 8. Wash twice in 1xPBS, 5 minutes each
[0068] 9. 10 μg / ml proteinase K in 1x proteinase K buffer for 5 min
[0069] 10. Wash twice in 1xPBS, 2 minutes each time
[0070] 11.4% PFA fixed on ice for 10 minutes
[0071] 12. Wash twice in 1xPBS, 5 minutes each
[0072] 13. Treat in 1xTEA [200ml of0,1M triethanolamine-HCl, pH8] containing 600μl acetic anhydride for 10 minutes
[0073] 14...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com