Haemophilus parasuis engineering strain with hhdA gene deletion and without resistance maker and construction method thereof
A technology of Haemophilus suis without resistance markers, which is applied in the field of construction of Haemophilus parasuis strains, and can solve the problems of unclear biological functions and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
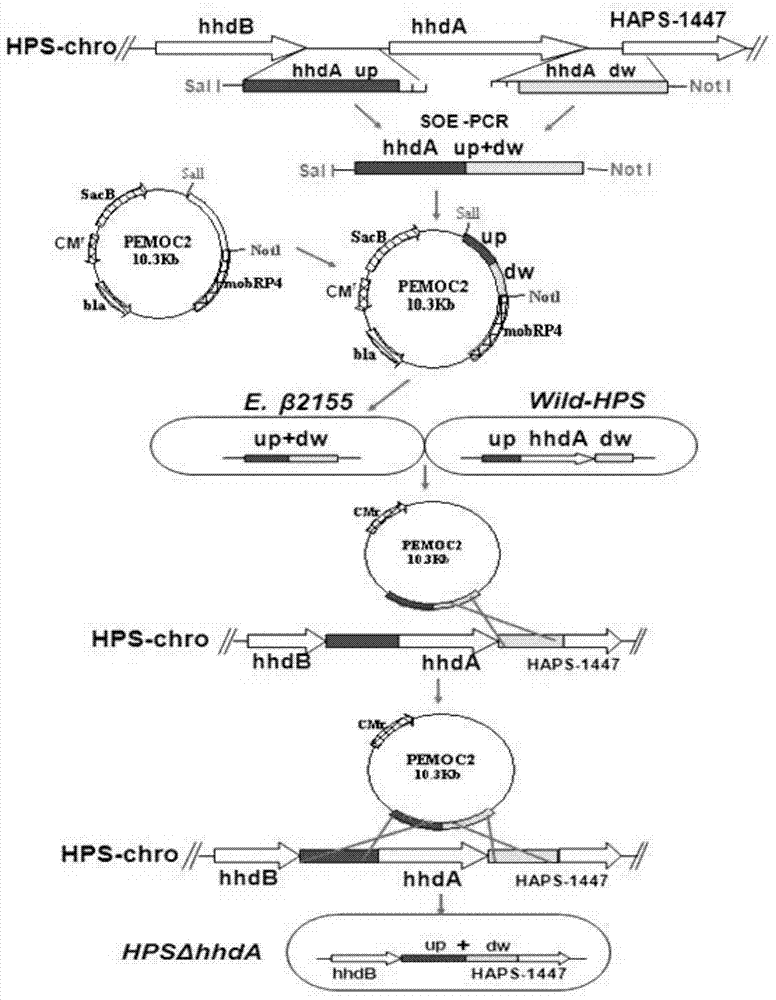
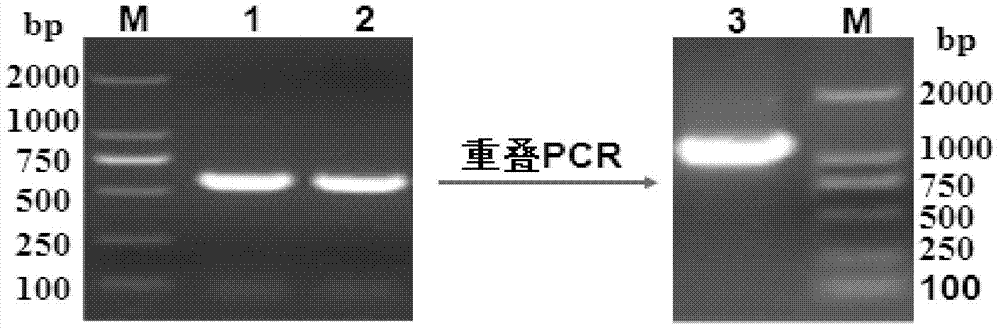
[0051] The embodiment uses the Haemophilus parasuis serotype 5 wild-type H11 bacterial strain ( Haemophilus parasuis HPS-H11) (hhdA +) (preserved in China Center for Type Culture Collection (CCTCC) on July 6, 2013, address: China, Wuhan, Wuhan University, deposit number is CCTCC NO: M 2013316) as the parent strain , respectively design specific primers for the upstream and downstream sequences of the hhdA gene, amplify the upstream and downstream homology arms, use the method of overlapping PCR to connect the upstream and downstream sequences, construct the suicide plasmid vector PEMOC2ΔhhdA, and transfer PEMOC2ΔhhdA into the parental strain by combining the transfer method Among them, the strain without resistance marker deletion of hhdA gene (HPSΔhhdA) was finally obtained after screening and identification by corresponding methods. Specific steps are as follows:
[0052] 1. Amplification of the upstream and downstream sequences of the HPS-hhdA gene
[0053] According ...
Embodiment 2
[0075] Identification of Genetic Stability of Gene Deletion in HPSΔhhdA Strain
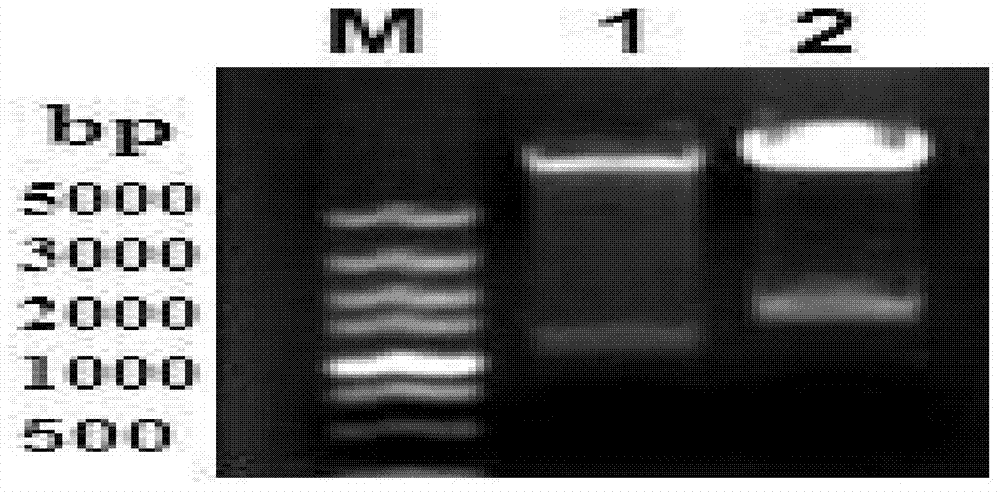
[0076] Streak culture the hhdA gene deletion strain HPSΔhhdA prepared in Example 1 on a TSA (10 μg / mL NAD, 10% fetal bovine serum) plate, pick a single colony and inoculate it on TSB (10 μg / mL NAD, 10% fetal bovine serum ) medium, cultured at 72°C and 200 r / min for 16 h, and the proliferated bacterial liquid was one generation. Then the first-generation bacterial solution was streaked on the TSA plate, and a single colony was picked and inoculated into the TSB medium for overnight shaking at 72°C. The proliferated bacterial solution was the second generation, which was repeatedly passaged in TSA and TSB medium. Continuously cultivated for 21 generations, every 3 generations, the proliferated bacterial liquid was amplified and identified by PCR with hhdA-up-F and hhdA-down-R primers, the amplification system was 25 μL: bacterial liquid template 1 μL, 10×PCR buffer 2.5 μL, 25 mmol / L MgCl 2 1 μL...
Embodiment 3
[0079] Identification of Growth Characteristics of HPSΔhhdA Strain
[0080] The hhdA gene-deleted strain HPSΔhhdA and the wild-type strain prepared in Example 1 were rejuvenated, respectively inoculated in TSB medium, cultured with shaking at 37 °C for about 12 h, and plate colonies were counted as seed liquid. According to the counting results, the deletion strain and the wild type strain were divided into 1×10 7 The colonies were inoculated into 5 mL of TSB liquid medium, and cultured with shaking at 37 °C 200 r / min, during which samples were taken every 2 h for plate colony counting, and the experimental results were drawn into a curve to analyze and identify the growth characteristics of the gene-deleted strains. Such as Figure 8 The indicated HPSΔhhdA strains had similar growth characteristics to the wild-type strain.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com