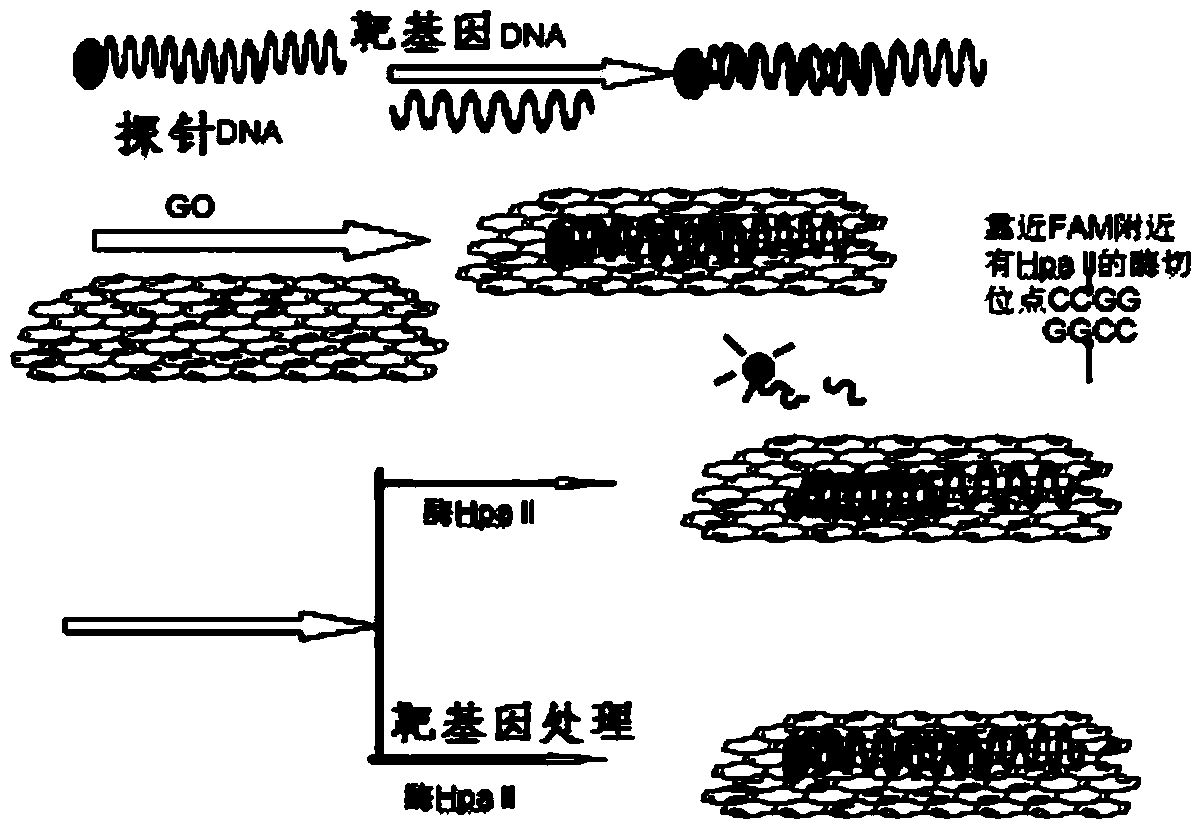
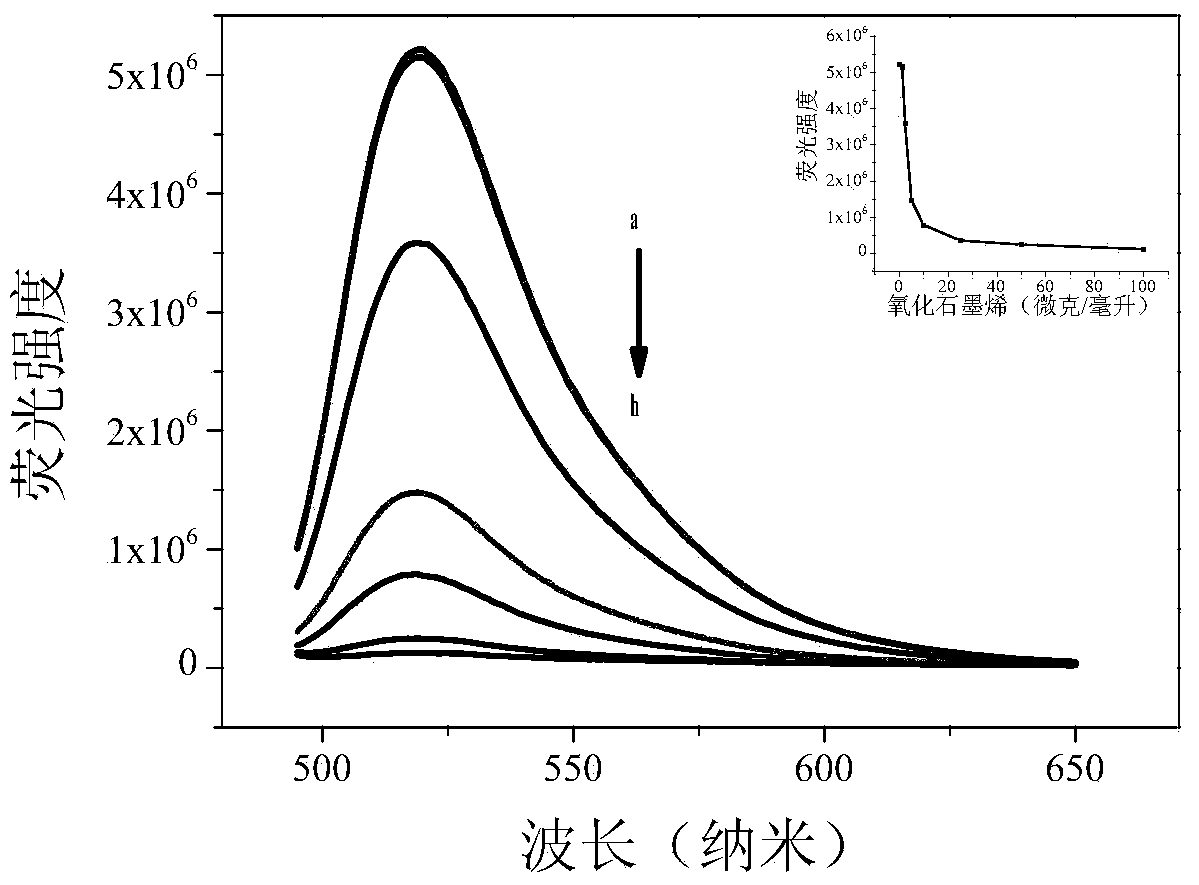
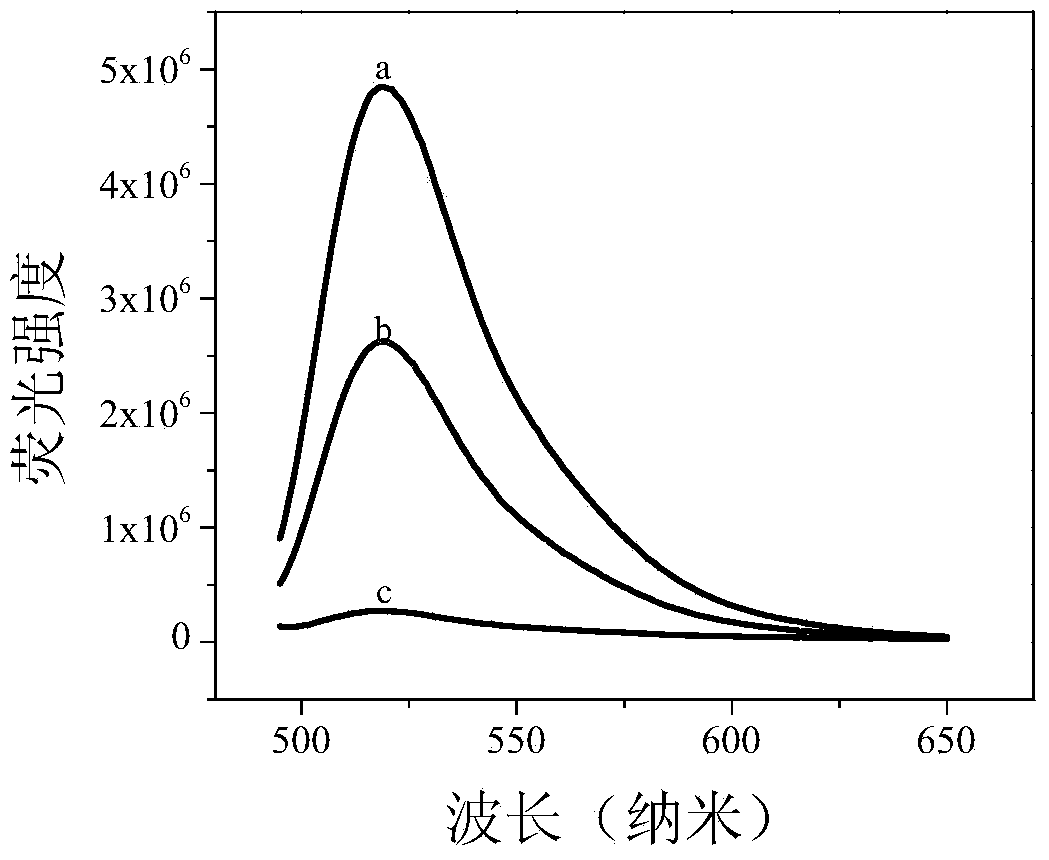
DNA methylation detection method based on graphene oxide and restriction enzyme and kit thereof
A technology of restriction endonuclease and detection method, which is applied in biochemical equipment and methods, microbial determination/inspection, material excitation analysis, etc., to achieve the effect of simple operation and simple and fast detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0048] Example 2 The process of using a detection kit to detect DNA methylation
[0049] DNA Methylation Detection Kit:
[0050] 1) 25 μg / ml graphene oxide;
[0051] 2) restriction endonuclease HpaII;
[0052] 3) Probe DNA containing a CCGG sequence modified with a fluorescent group at one end;
[0053] 4) There is a segment of target gene DNA complementary to the probe DNA.
[0054] 1. Detection of DNA methylation by methyltransferase:
[0055] In 33mM tris-acetic acid (10mM Mg 2+ , 66mM Na + ) buffer system with a concentration of 1 μM, hybridize the DNA sequence of the methylated target gene caused by the methyltransferase with the probe DNA modified with a fluorescent group at one end containing the CCGG sequence; gradually add 25 μg / ml Graphene oxide, and then add restriction endonuclease HpaII, observe the degree of fluorescence recovery after double-stranded DNA interacts with graphene oxide.
[0056] 2. Detection of DNA methylation caused by chemical reagents: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com