Method for building nucleotide formula to identify plant variety by using ribosome deoxyribonucleic acid (DNA)
A technology of nucleotide molecular formula and plant variety, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms, etc., can solve the problems of the same name of foreign objects, unclear source records, and restrictions on the production, promotion and breeding of crape myrtle varieties.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0113] Embodiment 1, the establishment and application of the nucleotide molecular formula identification method of Lagerstroemia indica varieties
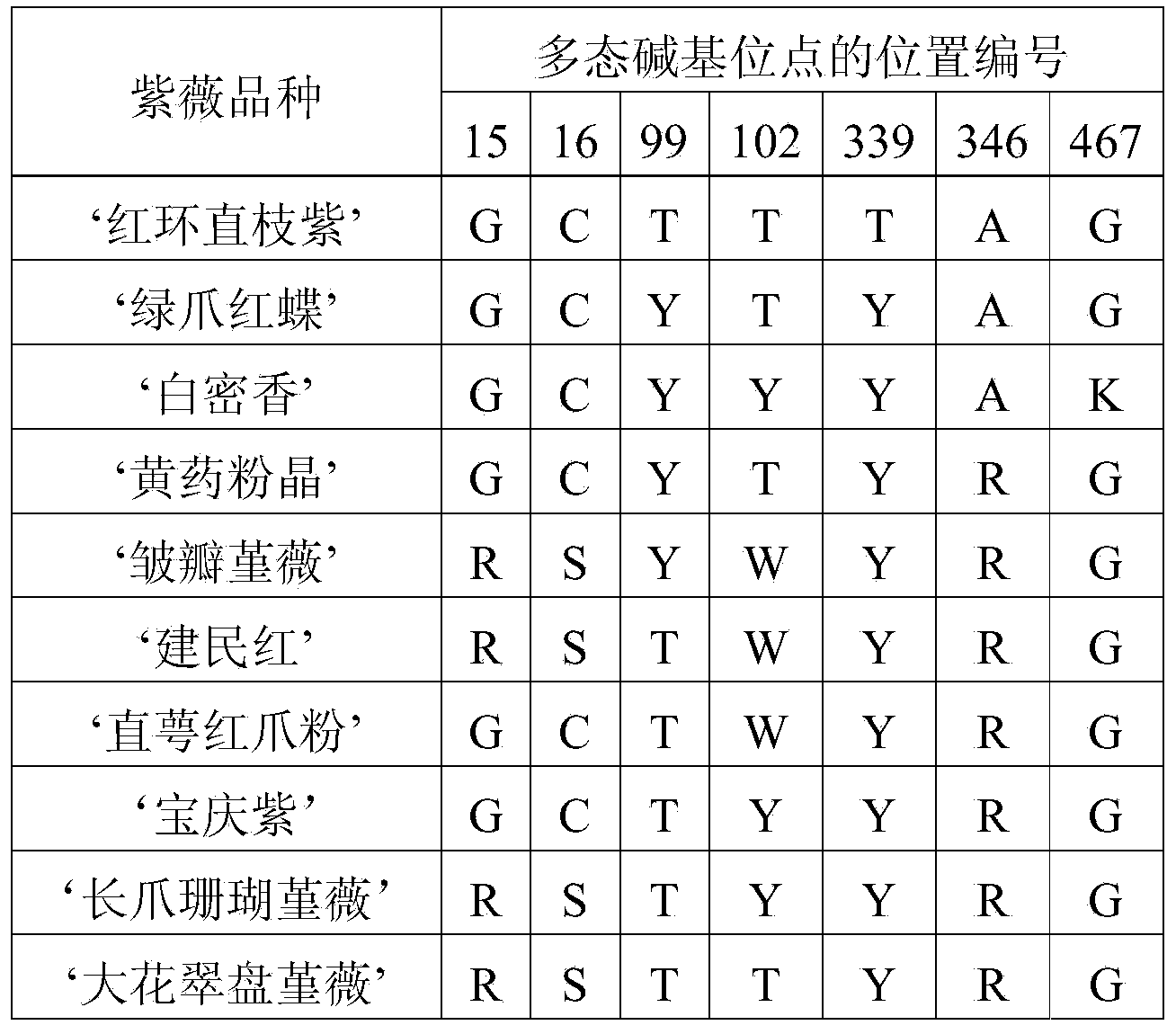
[0114] In order to construct a nucleotide molecular formula that can be used to effectively identify crape myrtle varieties, the inventors of the present invention obtained a DNA sequence containing a nuclear ribosomal DNA internal transcriptional spacer (ITS region) and part of the 26SrRNA gene from the following series of crape myrtle varieties, And the polymorphic sites among them were analyzed statistically. There are 10 kinds of crape myrtle varieties used as test materials, as follows: 'Honghuan Zhizhizi', 'Green Claw Red Butterfly', 'Bai Mixiang', 'Xantao Powder Crystal', 'Crinkle Petal Viola', 'Jianmin Red', 'Straight Calyx Red Claw Fen', 'Baoqing Purple', 'Long Claw Coral Viola' and 'Great Flower Viola Viola'.
[0115] 1. Primer design
[0116] According to the conserved regions at both ends of a region containing the I...
Embodiment 2
[0172] Embodiment 2, identify other crape myrtle samples
[0173] In order to verify the specificity of the nucleotide molecular formula established in Example 1 to the Lagerstroemia indica plant material, the inventors of the present invention also collected the leaves of Lagerstroemia indica, and extracted its genomic DNA according to the method in Example 1, and used sequence 1 and The primer pair composed of two single-stranded DNA molecules shown in sequence 2 was amplified by PCR and sequenced, and the zw_ITS sequence of Lagerstroemia indica was obtained (sequence 13, compared with the zw_ITS sequences of 10 crape myrtle varieties described in Example 1 , it was found that the 108th nucleotide of Lagerstroemia nadina was missing). Put together the ITS sequence of Lagerstroemia indica and the sequences of the aforementioned 10 varieties of Lagerstroemia indica and compare them using software (such as Mega5 or ClustalX) to obtain a sequence alignment matrix (alignment), an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com