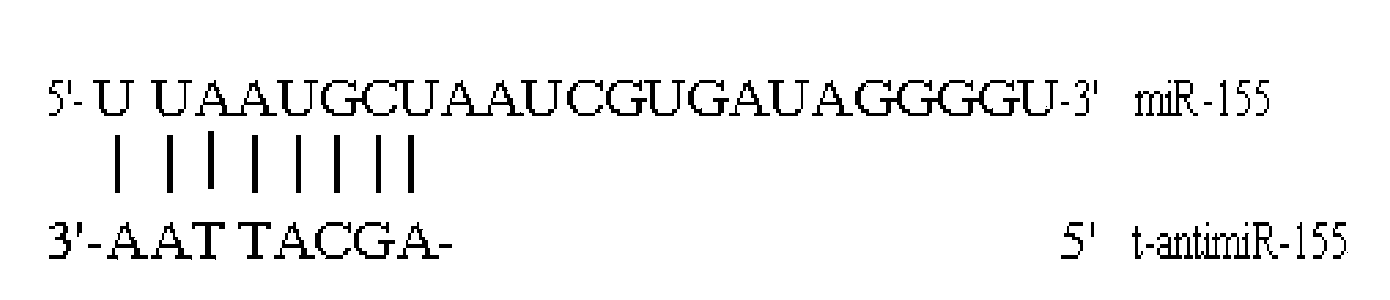Antisense oligodeoxynucleotide for micro RNA-155 seed sequence and application
A technology of RNA-155 and antisense oligonucleotides, applied in the field of antisense oligonucleotides, can solve the problems of G1 phase arrest and unreported effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1. Experimental materials and methods
[0028] 1. Design and synthesis of antisense oligonucleotides
[0029] The seed sequence of human microRNA-155 was obtained from the microRNA Families database, and the antisense oligonucleotide sequence (t-antimiR-155) targeting the seed sequence was designed according to the principle of sequence complementarity, and the antisense oligonucleotide sequence was determined by BLAST software analysis Acid sequences and random control sequences (such as figure 1 shown).
[0030] 2. Main reagents
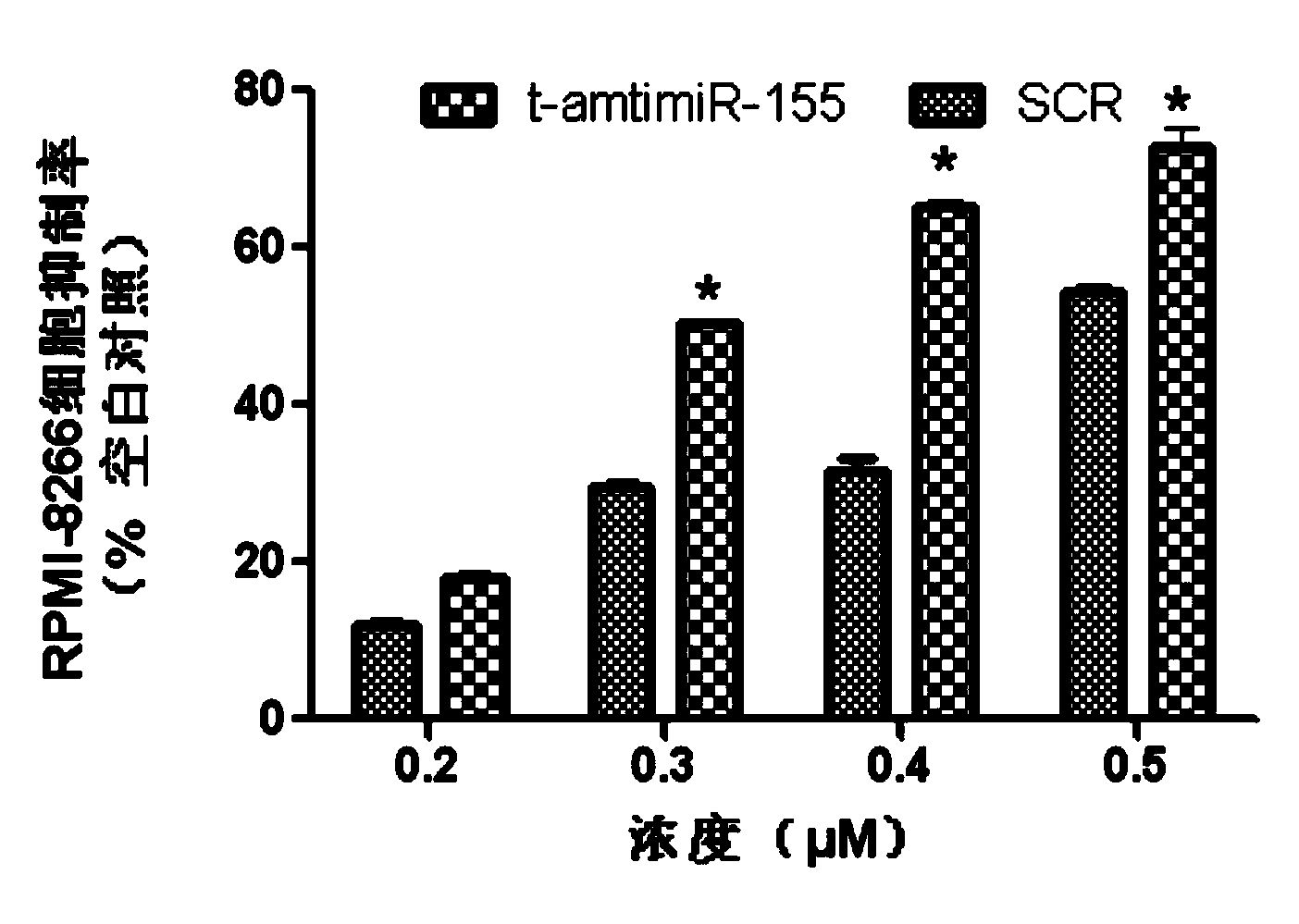
[0031] MTT (tetramethylazolazolium blue) and DMSO (dimethyl sulfoxide) were purchased from Sigma; fetal bovine serum and RPMI-1640 medium were purchased from GIBCO; Lipofectamine TM 2000 was purchased from Invitrogen. RPMI-8266 cell line was purchased from Shanghai Baili Biotechnology Co., Ltd. Antisense oligonucleotide sequence (t-antimiR-155): 5′-AGCATTAA-3′; random sequence (scramble, SCR):
[0032] 5′-TCATACTA-3′, all synthesized ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com