A molecular detection kit for Chestnut Phytophthora and its application method
A chestnut pathogen and molecular detection technology, applied in the field of microbiology, can solve the problems of inability to achieve rapid and accurate detection and identification of pathogenic bacteria, difficult to achieve accurate identification of similarity, false positives, etc., to achieve rapid and accurate early diagnosis and overcome poor stability , the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0027] The main reagents used: DNA extraction kit and recovery kit were purchased from Tiangen Biochemical Technology Company, DH5αE.coli Competent and Premix were purchased from TaKaRa Company, primer synthesis and PCR product sequencing were completed by Shanghai Sangon Bioengineering Company.
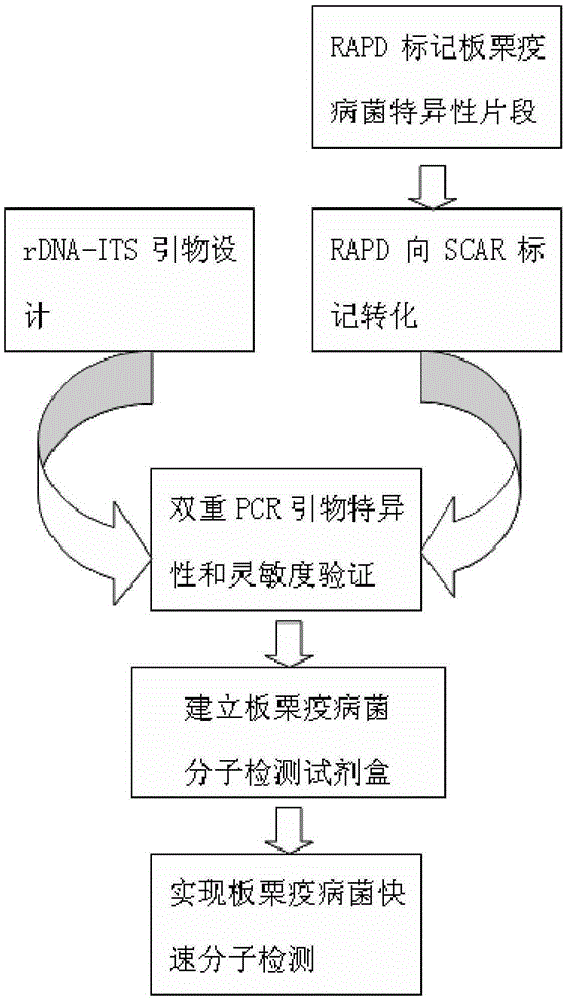
[0028] figure 1 It is the implementation diagram of the dual PCR detection technology for Phytophthora chestnut.
[0029] 1. Design of double PCR primers for rapid molecular detection of Phytophthora chestnut
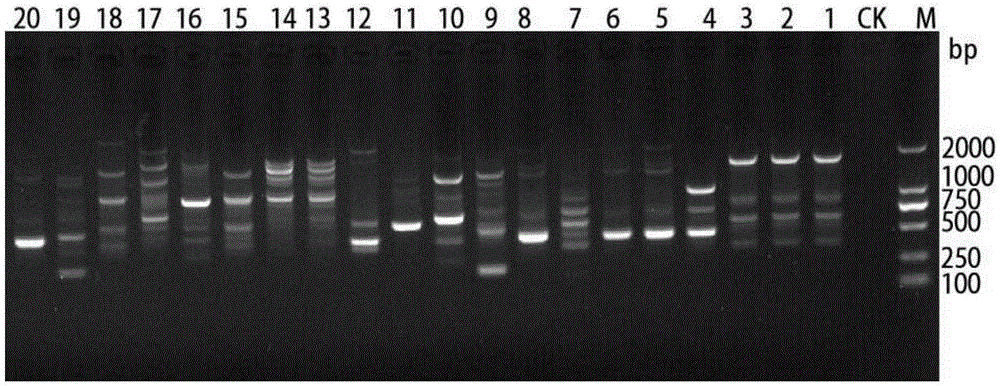
[0030](1) Primers in the rDNA-ITS region: Purify and cultivate the Chestnut Phytophthora bacterium isolated from chestnut cultivation areas in Chongqing, Sichuan, Ya'an, Sichuan, and Luzhou, Sichuan. First, inoculate the tested strain on PDA medium, and place it in a dark condition in an incubator at 25°C Cultivate under low temperature for 3 days, then pick 3~4 pieces of bacteria cakes about 5mm from the edge of the colony, inoculate them in a conical flask containing 100ml ...
Embodiment 2
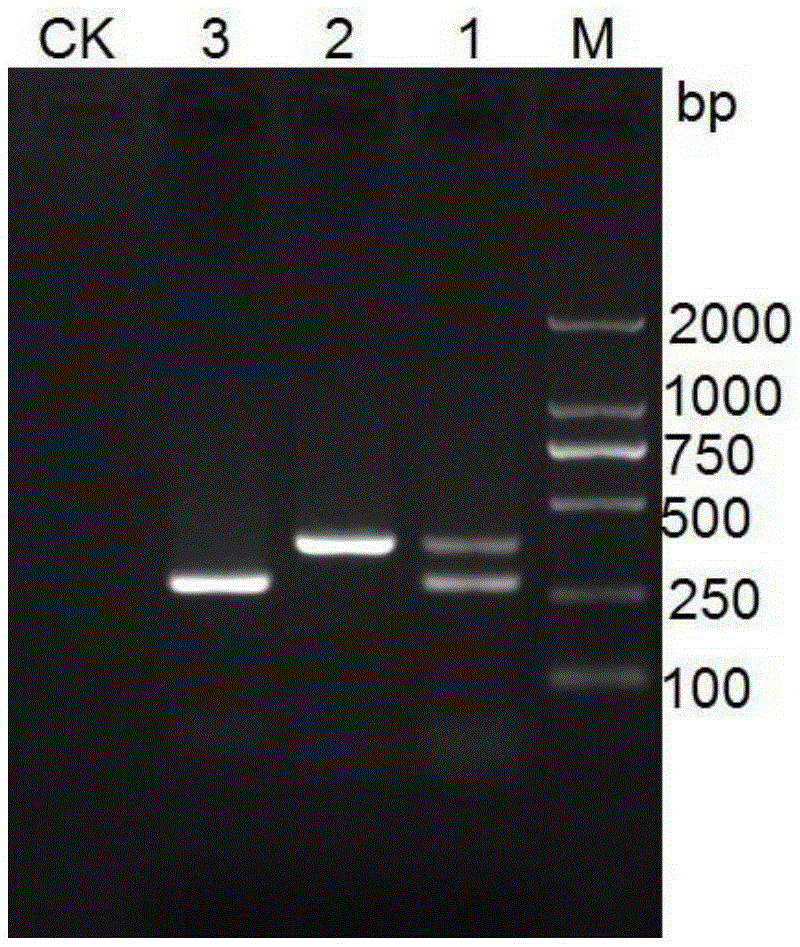
[0091] Example 2 Detecting Field Diseased Plants with Chestnut Phytophthora Molecular Detection Kit
[0092] 1. Genomic DNA extraction:
[0093] Genomic DNA was extracted by the CTAB method as follows:
[0094] 1) Cut out a section of 50-100 mg suspected chestnut blight disease tissue with a scalpel, disinfect the surface with 70% alcohol, rinse it under distilled water, dry the water with absorbent paper, transfer it to a mortar and grind it quickly with liquid nitrogen (for easy Grinding can be properly cut into powder, put into a 1.5mL centrifuge tube;
[0095] 2) Add 900 μL of CTAB solution and 90 μL of 10% SDS after preheating at 60°C, and mix well;
[0096] 3) 60°C water bath for 1 hour, in the middle every 10mm upside down and mix once;
[0097] 4) Centrifuge at 12000rpm for 10min;
[0098] 5) Add 700 μL volume of phenol:chloroform:isoamyl alcohol (25:24:1) to the supernatant, mix well, and let stand for 10 minutes;
[0099] 6) Centrifuge at 12000rpm for 10min;
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com