Molecular marker closely linked with corn bacterial wilt resistance genes and primer and application thereof
A molecular marker and resistance gene technology, applied in the field of agricultural biology, can solve the problems of low selection efficiency and long breeding cycle, and achieve the effect of reducing labor land, shortening breeding cycle, and improving corn yield level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1, Obtaining the special primer pair for the identification of bacterial wilt resistance traits: the nucleotide sequence shown in SEQ ID NO: 3 in the sequence table and the nucleotide sequence shown in SEQ ID NO: 4 in the sequence table
[0029] 1. Parental Genome Amplification
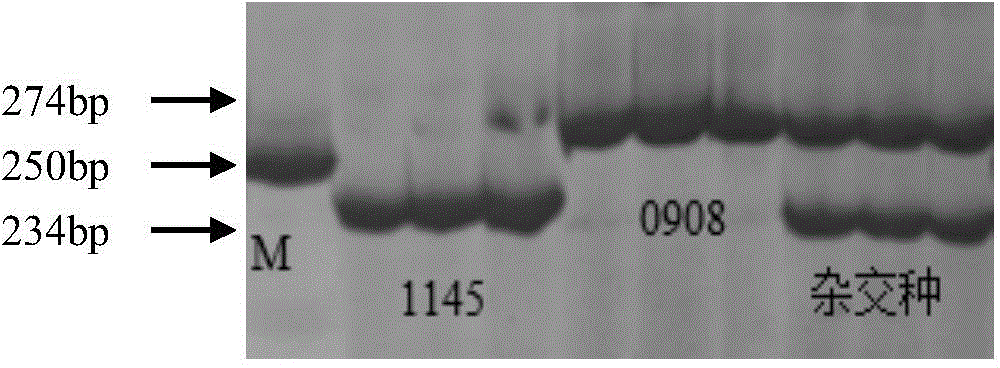
[0030] The parents used to create the maize recombinant inbred line (RIL) population are 1145 and 0908 selected by our company. 0908 is the female parent, which is not resistant to bacterial wilt; 1145 is the male parent, which is highly resistant to bacterial wilt.
[0031]Genomic DNA of parental leaves was extracted by CTAB method, using primers published by Qing Yang (Qing Yang, Zhi Li, et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. PNAS 2013, 10.) Yes, it is composed of the nucleotide sequence shown in SEQ ID NO: 5 in the sequence table and the nucleotide sequence shown in SEQ ID NO: 6 in the ...
Embodiment 2
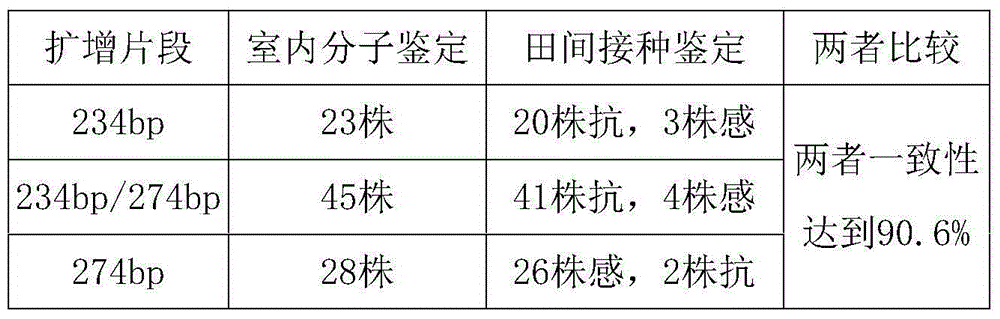
[0038] Example 2, Correlation verification between the amplification product of QK1 and the resistance traits of corn bacterial wilt
[0039] 96 F 2 The substitute material is the test object. Genomic DNA of corn endosperm was extracted by alkaline boiling method, and the specific steps were the same as in Example 1. PCR amplification experiments were performed with QK1. The PCR reaction system is: the PCR reaction system is: 10X Buffer 2 μ l (containing Mg2+), dNTP0.4 μ l (10 mM), the nucleotide sequence shown in the sequence table SEQ ID NO: 3 and the sequence table SEQ ID NO: shown in 4 2 μl of each nucleotide sequence (5 μM), 1 μl of genomic DNA extracted from corn to be tested, 0.5 μl of taq enzyme (5 U / μl), the reaction volume is 20 μl, and a drop of mineral oil is added to cover. The PCR reaction program was: 94°C 5min; 94°C 60s, 53.5°C 60s, 72°C 60s, 35 cycles; 72°C 10min.
[0040] The PCR amplified products were analyzed by 6% polyacrylamide gel electrophoresis, s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com