Glaucoma screening kit
A kit, glaucoma technology, applied in the field of SNP
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Sample Collection and Genomic DNA Extraction
[0057]The primary open-angle glaucoma included in the study of the present invention includes high intraocular tension glaucoma (HTG) and normal intraocular tension glaucoma (NPG). The discovery phase of the genome-wide association analysis (GWAS), the validation phase 1 and the validation phase 2 were all used for ocular hypertension glaucoma (HTG). The discovery phase included 1007 sporadic ocular hypertension glaucoma patients and 1009 normal controls. All samples were collected from Shanghai, Hong Kong, and Chengdu, respectively in the Eye, Ear, Nose and Throat Hospital Affiliated to Fudan University, the Hong Kong Eye Hospital of the Chinese University of Hong Kong, and the Sichuan Provincial People's Hospital. The first stage of validation included 525 patients with ocular hypertension glaucoma and 912 normal controls. The samples were taken from the Singapore National Eye Center, Singapore Eye Research Ins...
Embodiment 2
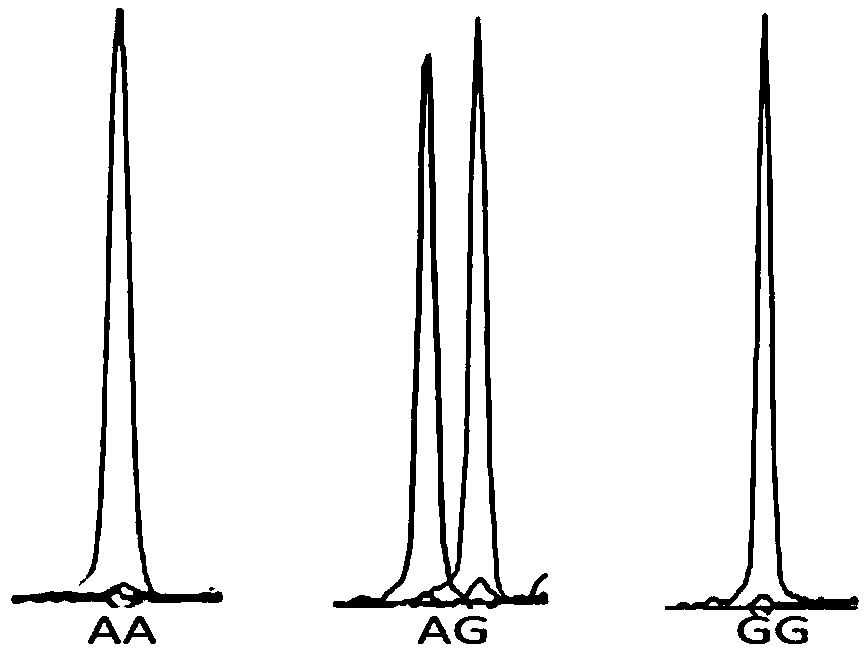
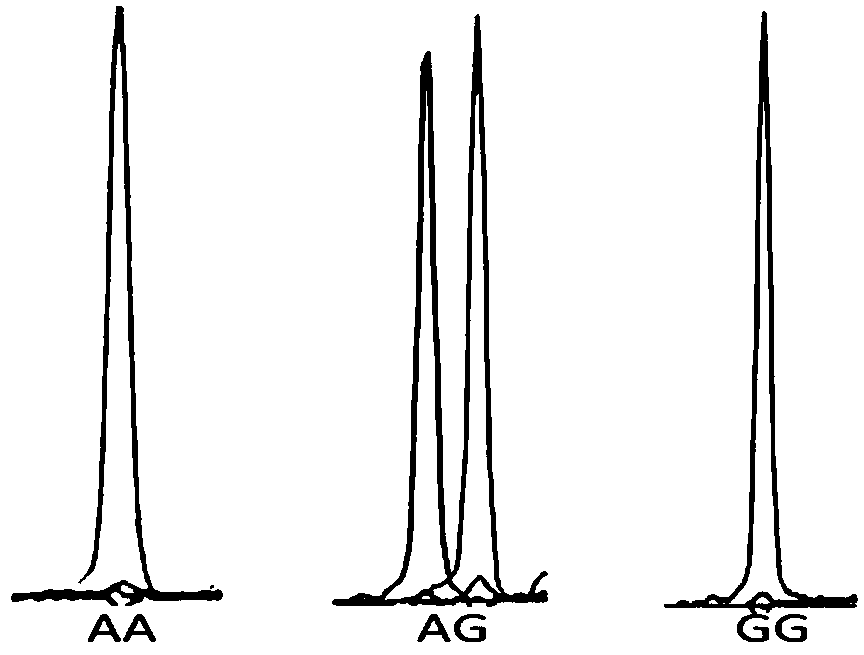
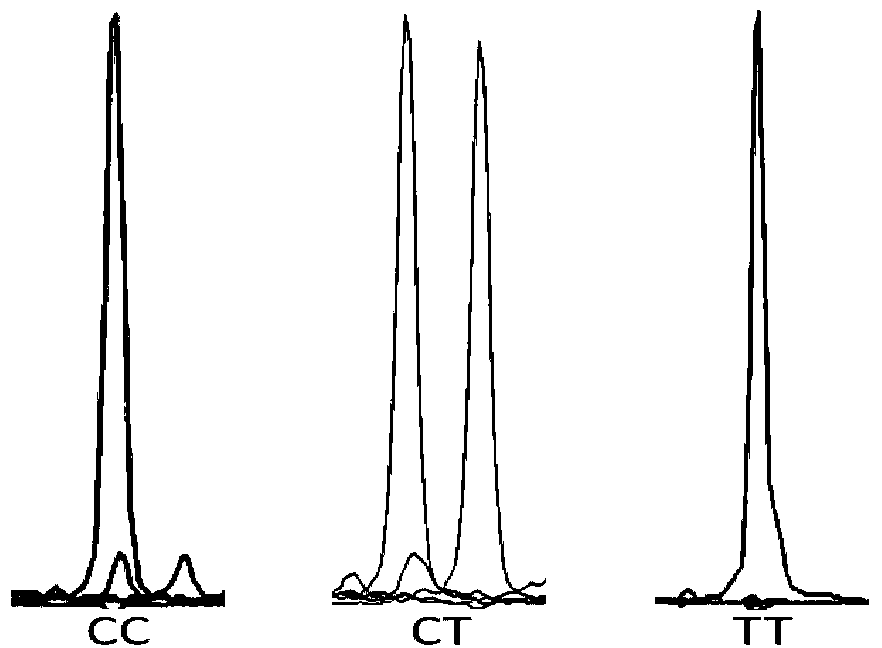
[0059] Embodiment 2 Genotype analysis of SNP of the present invention
[0060] The present invention uses the Snapshot genotyping technology to detect the following 31 SNP sites located on the ATP-binding cassette, sub-family A (ABC1), member1, ABCA1) gene. Gene amplification reagents and Snapshot primers were used for typing according to the following methods. SNaPshot is also called micro-sequencing method. This technology, developed by Applied Biotechnology (ABI), is a typing technology based on the principle of fluorescently labeled single base extension, and is mainly aimed at medium-throughput SNP typing projects. In a reaction system containing sequencing enzymes, four fluorescently labeled ddNTPs, different length extension primers close to the 5' end of the polymorphic site, and PCR product templates, the primers are terminated after one base extension, and after electrophoresis on the ABI sequencer, According to the color of the peak, the type of base incorporated ...
Embodiment 3
[0232] The genotype analysis of embodiment 3 SNP of the present invention
[0233] Except the Snapshot detection method that embodiment 2 enumerates, 31 SNPs of the present invention can also adopt following detection method to detect:
[0234] SNP detection methods include:
[0235] 1. DNA sequencing technology:
[0236] 1. The dideoxynucleotide chain termination method (traditional Sanger sequencing) is based on the fact that the nucleotide starts at a fixed point and terminates at a specific base at random, and fluorescent labels are carried out after each base. Produce a series of four groups of nucleotides of different lengths ending with A, T, C, G, and then detect by electrophoresis on urea-denatured PAGE gel, so as to obtain the visible DNA base sequence. Basic process: 1), DNA template extraction and quality inspection; 2), sequence alignment, primer design and synthesis; 3), PCR amplification, including preparation of denatured double-stranded templates, extension ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com