A kind of dna ligase activity assay method
A DNA ligase and activity determination technology, applied in the field of biochemistry, can solve the problems of low experimental cost, complex probe design, unsatisfactory, etc., and achieve the effect of simple operation steps, easy primer design, and fast operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1. Feasibility verification of T4 DNA ligase activity by "transformation method"
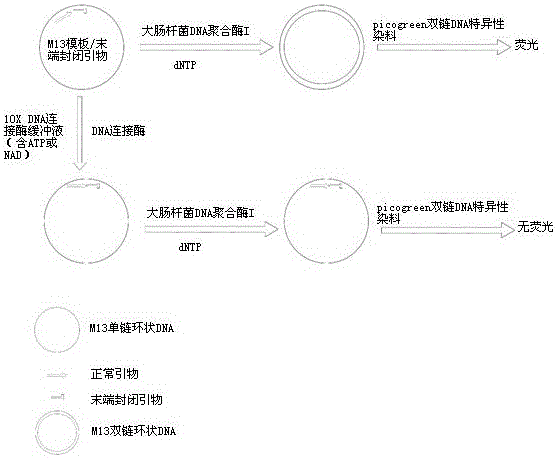
[0024] Preparation of M13 template / normal primer-3' terminal phosphorylated primer complex:
[0025] M13 single-stranded DNA: M13mp18 single-stranded DNA (NEB);
[0026] Normal M13 primer: 5'-aagccatccgcaaaaatgacctct-3';
[0027] 3'-terminal phosphorylated M13 primer: 5'-tatcaaaaggagcaattaaagg-3' (3'-terminal hydroxyl group is phosphorylated)
[0028] M13 template / normal primer-3'-terminal phosphorylated primer complex: M13mp18 single-stranded DNA was mixed with normal M13 primer, 3'-terminal phosphorylated M13 primer in a molar ratio of 1:1:1, in annealing buffer (10 mM Tris -HCl pH 8.0, 50 mM NaCl), after heating at 70 °C for 5 min, slowly cool to room temperature to anneal the template primers.
[0029] Preparation of M13 template / 3' terminal phosphorylated primer complexes:
[0030] M13 single-stranded DNA: M13mp18 single-stranded DNA (NEB);
[0031] 3'-terminal phosphory...
Embodiment 2
[0050] Example 2. Drawing of T4 DNA ligase activity-fluorescence intensity curve and EC50 calculation
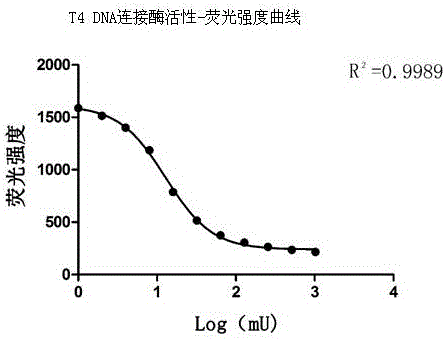
[0051] Take the log value of the T4 DNA ligase activity unit (mU) of 2#-12# as the abscissa and the fluorescence intensity as the ordinate, and use GraphPad Prism5 to make a nonlinear regression curve. We found that these data points can be well fitted to an inverted sigmoid curve, such as figure 2 shown.
[0052] By repeating the experiment multiple times and calculating the half-maximal effect concentration (EC50) of the curve, we found that the EC50 value remained stable across experiments:
[0053] experiment
[0054] Average (mU)
Embodiment 3
[0055] Example 3. Drawing of DNA ligase activity-fluorescence intensity standard curve from different sources and definition of "transformation method" activity
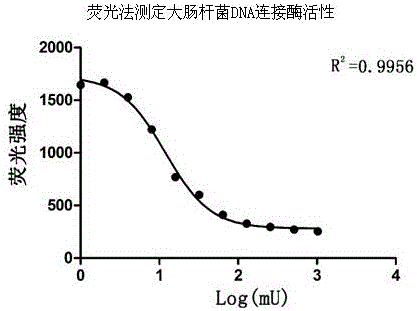
[0056] We also selected a different species of DNA ligase, namely E. coli DNA ligase (enzymaticsL6090L), and according to the methods of Example 1 and Example 2, the activity-fluorescence intensity standard curve was drawn and EC50 was calculated. All DNA ligase activities were determined using standard semi-quantitative assays. The unit commonly used to describe ligase activity is cohesive end unit (CEU): 1 cohesive end activity unit refers to 50% of the ligase digested by HindIII in 20μL of 1×T4 DNA ligation reaction buffer at 16°C for 30min. The amount of enzyme required for ligation of the λ DNA fragment [5' end at a concentration of 0.12 μM (300 μg / mL)]. The result is as image 3 shown.
[0057] species
source
EC50 (mU)
T4 phage
NEB E. coli recombinant expression
12.84±0.17*
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com