Method for developing genome simple sequence repeats (SSR) molecular marker
A molecular marker and genome technology, applied in the field of molecular biology, can solve the problems of difficult operation, primer-specific reading errors, time-consuming, etc., and achieve the effect of improving efficiency, reducing human and financial resources, and reducing sequencing costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
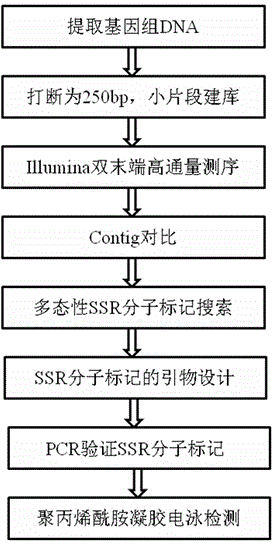
[0032] Materials: Colocasia gigantea was selected as the test sample, which has no genome and transcriptome except a few chloroplast genes in GeneBank. Take two wild taro plants, one from Xishuangbanna, Yunnan Province, China, and the other from Vietnam. See flow chart figure 1 .
[0033] method:
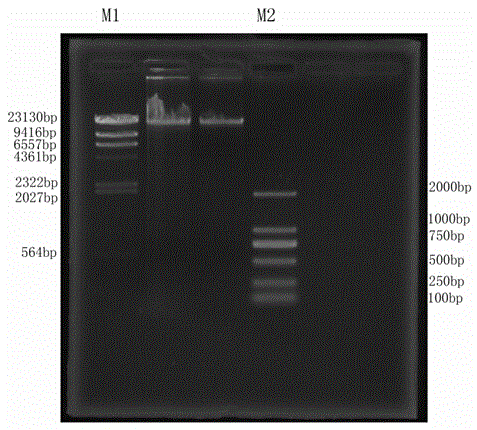
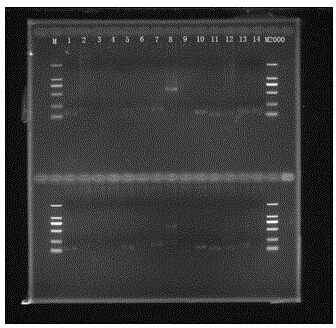
[0034] (1) DNA extraction: cut 0.4 g of fresh and tender leaves and extract DNA with the CTAB method, and then use agarose gel electrophoresis to detect the extraction effect, such as figure 2 As shown in , samples with clear main band and no smear can be used for subsequent library construction ope...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com