DNA (Deoxyribonucleic Acid) library for detecting cholestatic jaundice pathogenic genes and application thereof
A DNA library, cholestasis technology, applied in the field of DNA library, can solve the problems of indistinguishable, unable to meet the requirements of multi-sample detection timeliness, low throughput, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0054] 1. Reagents used in the method:
[0055] Ion AmpliSeq TM Library Kit 2.0, Ion PGM TM Template OT2200Kit v3, Ion Sequencing 200Kit v2, Ion Xpress Barcode Adapters 1-16Kit, Ion 318 TM Chip Kit v2
[0056] 2. Specimen collection and storage
[0057] (1) Specimen collection: The specimen is the peripheral blood of the patient. Blood is 5ml of venous blood taken routinely, treated with EDTA anticoagulant.
[0058] (2) Storage: It can be detected immediately, stored at 4°C for one week, and stored at -80°C for more than one week.
[0059] 3. Detection steps and result analysis:
[0060] (1) Extraction of genomic DNA of specimens: DNA extraction of specimens was carried out according to the operation instructions of the blood DNA extraction kit of Tiangen Biochemical Technology (Beijing) Co., Ltd.
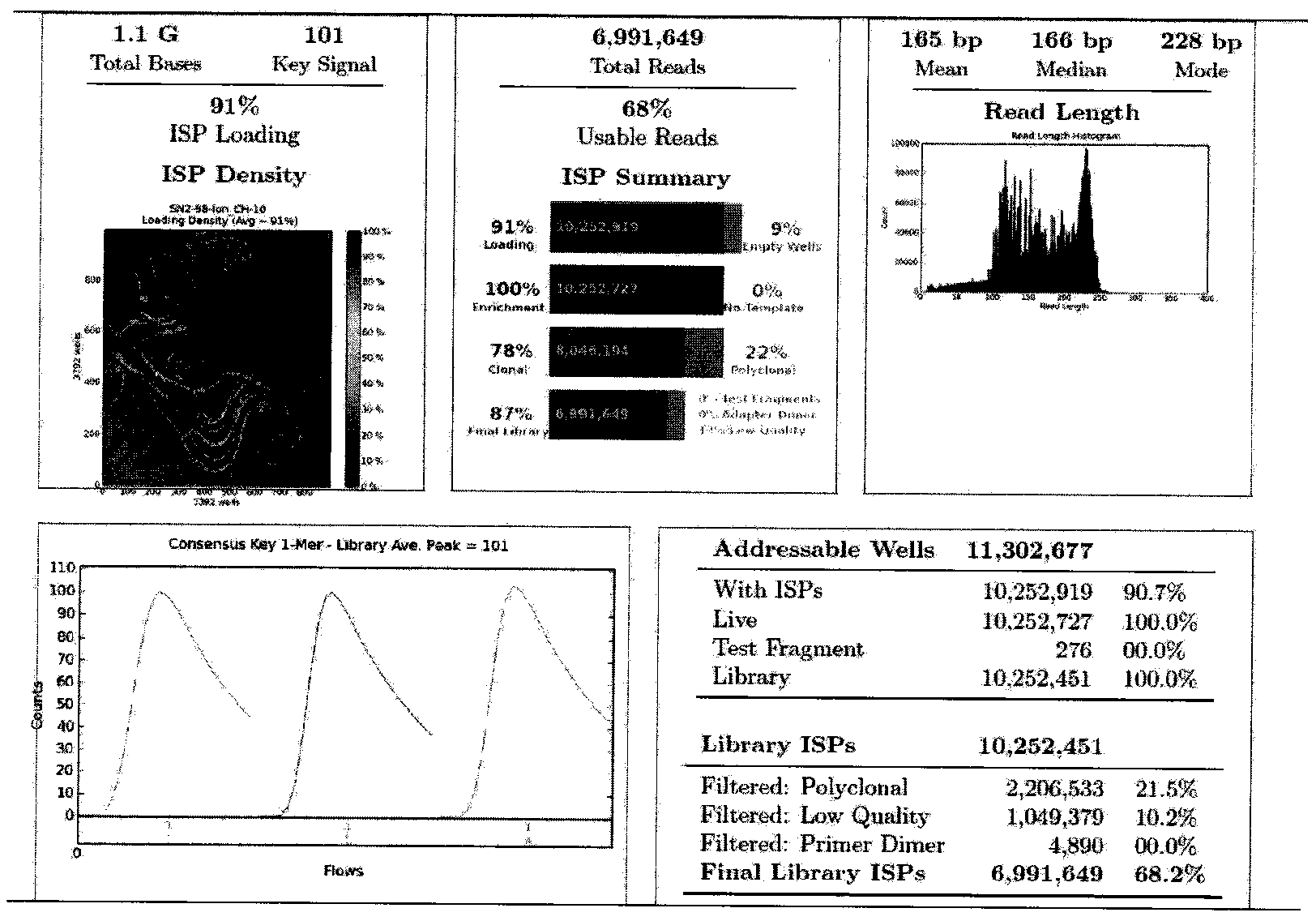
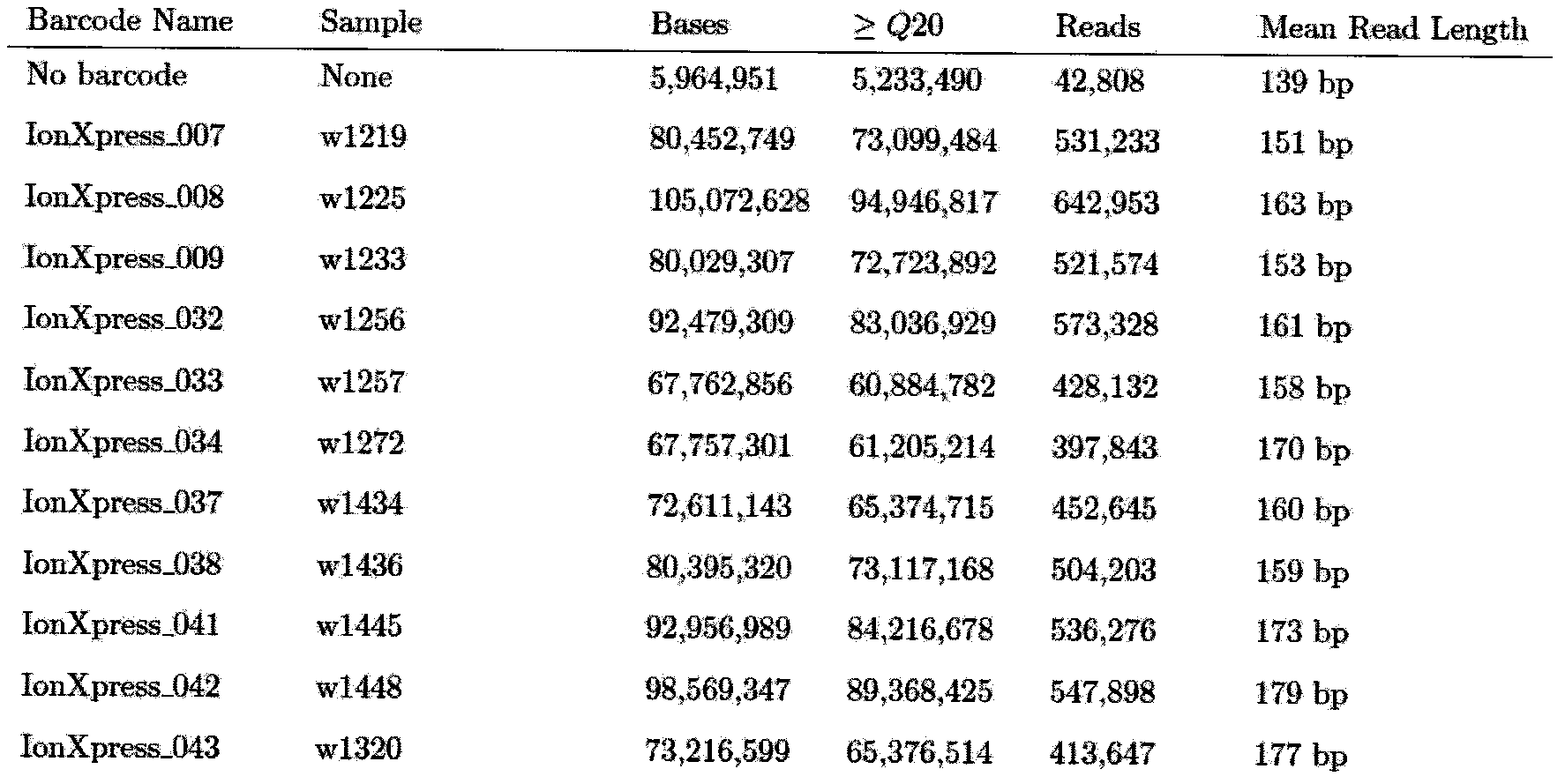
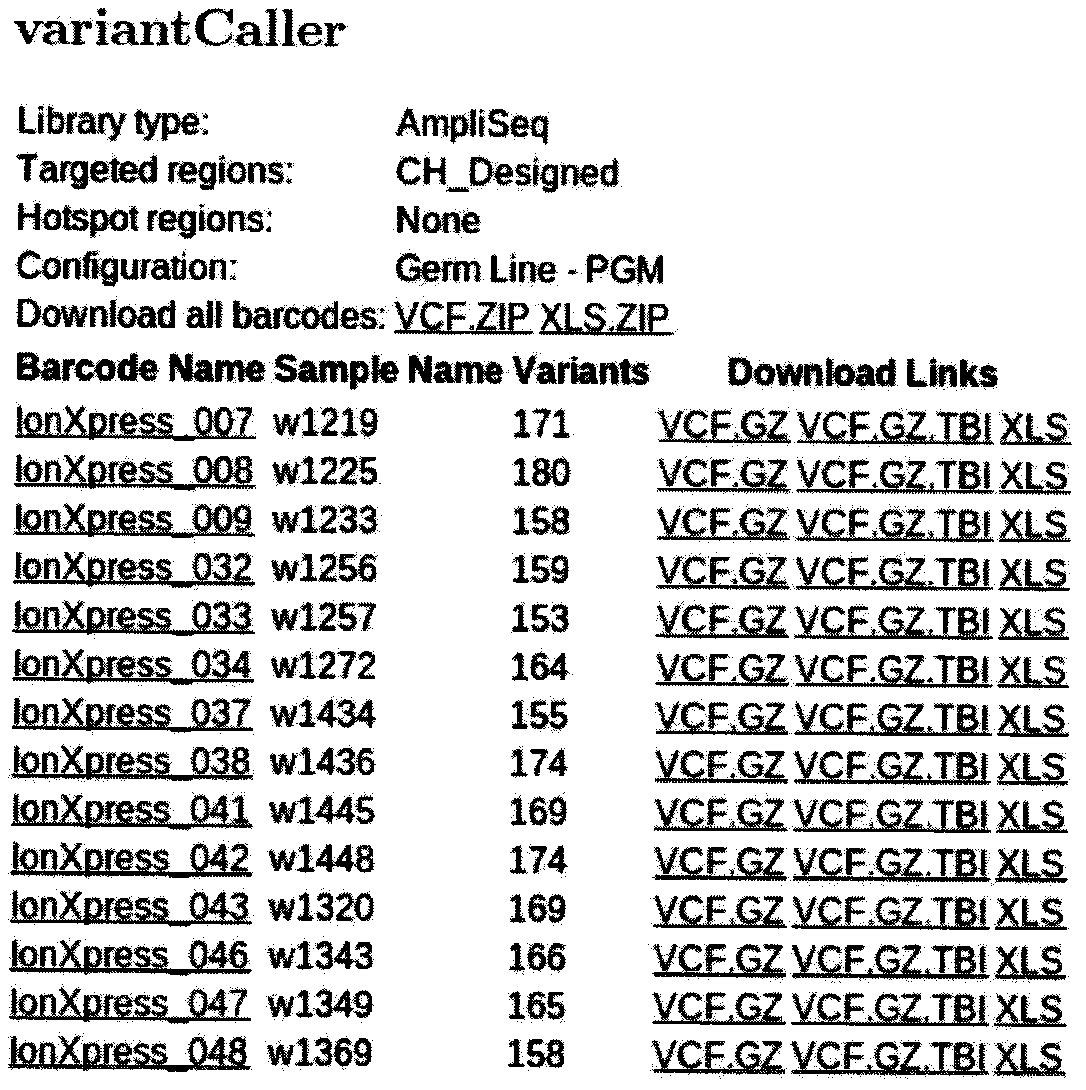
[0061] (2) Super multiplex PCR amplification and library building of the target detection region: the whole exons of 60 genes involved in the present invention are used...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com