Serotyping PCR (polymerase chain reaction) method for haemophilus parasuis
A technology for Haemophilus suis and serotyping, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection. It can solve problems such as backwardness and impracticality, and achieve high accuracy and short time consumption. , good specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Design of PCR primers
[0042] Compare the conserved genes on the CPS gene cluster of Haemophilus parasuis, and analyze and compare the differences in the conserved genes among the serotypes, design multiple pairs of primers for each serotype, and compare their specificity, sensitivity and other factors through a large number of experiments Finally, a pair of primers with good specificity and high sensitivity are determined for each serotype as identification PCR primers. The sequences of the PCR primers are shown in Table 1, and the sequences of the PCR primers are shown in SEQ ID NO.1-SEQ ID NO.30.
[0043] Table 1 The sequences of PCR primers for each serotype
[0044]
[0045]
Embodiment 2
[0046] Embodiment 2: the establishment of serotyping PCR method
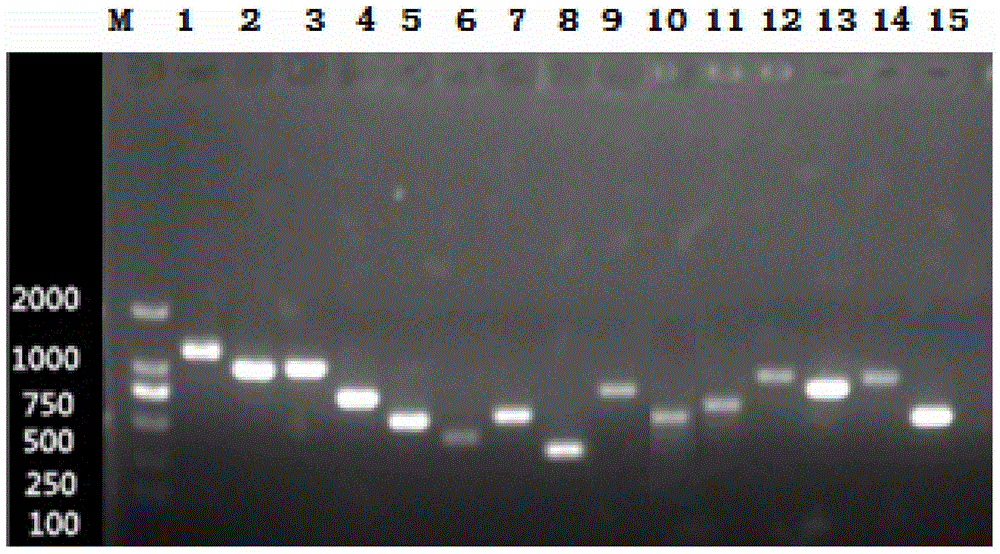
[0047] PCR amplification: Using the genomic DNA of the reference strains of 15 serotypes of Haemophilus parasuis as a template and sterile double-distilled water as a negative control, perform PCR amplification on the 15 pairs of primers designed above.
[0048] All PCR amplifications use a 50 μL reaction system: 25 μL of 2×Taq PCR Master Mix, 1 μL of each primer (10 mmol / mL), 1 μL of template DNA, and then add sterile double distilled water or ultrapure water to make up to 50 μL.
[0049] The PCR reaction procedure is as follows: the system is pre-denatured first, and then undergoes cycles of denaturation-annealing-extension in sequence. Wherein, the pre-denaturation temperature is 95° C., and the pre-denaturation is 5 minutes. In the cycle, the denaturation temperature is 94°C, denaturation is 45s, annealing temperature is 59°C, annealing is 30s, extension temperature is 72°C, extension is 30s, and the numb...
Embodiment 3
[0051] Embodiment 3: the specificity test of serotyping PCR method
[0052] In order to identify the specificity of the serotyping PCR method provided by the present invention to a single serotype, the typing primers of each Haemophilus parasuis were subjected to PCR reaction with the genomic DNA of other 14 serotype strains. Test results such as Figure 2-16 As shown, the above results confirmed that the various types of PCR primers did not cross-react with the other 14 serotypes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com