Nanobody against deoxynivalenol antibody
A deoxynivalenol, nanobody technology, applied in biochemical equipment and methods, applications, instruments, etc., can solve the problems that restrict the application and promotion of immunological detection methods, the health and environmental threats of detection personnel, and the high price. problem, to achieve good effect, cost saving, harm reduction effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Construction of camel-derived natural single domain heavy chain antibody library
[0027] 1) Separation of camel-derived leukocytes: add lymphocyte separation solution to the centrifuge tube, then slowly add an equal volume of blood sample, centrifuge at 1000 g for 50 min; carefully draw the leukocytes suspended in the middle layer into a new centrifuge tube, add 1 / 2 volumes of PBS, centrifuge at 1000g for 15min; discard the supernatant, wash the leukocytes on the wall of the centrifuge tube with PBS, centrifuge at 1000g for 10min; discard the supernatant, add 500 μL PBS to resuspend the leukocytes and count; add at a volume ratio of 1:15 The lysate (RNAiso) was saved for future use.
[0028]2) Extraction of total RNA: add 1 / 4 volume of chloroform to the above lysate, shake vigorously for 20 s to fully emulsify, let stand at room temperature for 5 min; centrifuge at 12000g at 4°C for 15 min, transfer the supernatant to another fresh centrifuge Add an equal ...
Embodiment 2
[0050] Example 2. Affinity panning and identification of Nanobodies
[0051] 1) Affinity panning of Nanobodies: First, dilute the anti-DON monoclonal antibody with PBS (pH 7.4) to a final concentration of 100 μg / mL, and coat at 4°C overnight. The next day, after washing 5 times with PBST (10mM PBS, 0.1% Tween-20 (v / v) ), add 5% BSA-PBS (or 5% OVA-PBS) to block for 1 hour at 37°C. Then wash 6 times with PBST, add 100 μL camel-derived natural single domain heavy chain antibody library (titer about 2.0×10 11 cfu), incubate at 37°C for 2 hours. Unbound phages were discarded, washed 10 times with PBST, added 100 μL of Glycine-HCl (0.2M, pH 2.2) to elute for 8 min, and immediately neutralized with 15 μL of Tris-HCl (1M, pH 9.1). Take 10 μL of the eluted phage to determine the titer, and the rest is used to infect 25 mL of E. coli The TG1 strain was amplified. On the third day, the amplified phage was precipitated with PEG / NaCl, and the titer of the phage was determined.
[0...
Embodiment 3
[0055] Example 3. Sequencing of Nanobody Encoding Gene and Determination of its Amino Acid Sequence
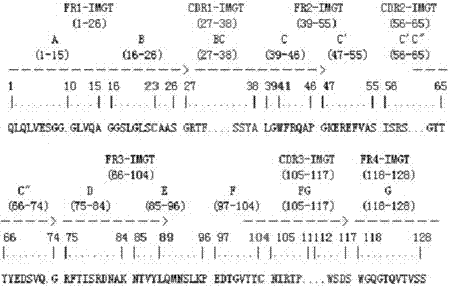
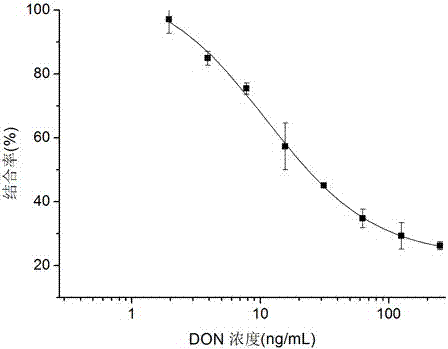
[0056] The phage clones displaying positive Nanobodies identified by indirect competition ELISA were subjected to DNA sequencing, and the amino acid sequence of the Nanobodies could be obtained according to the DNA sequencing results and the codon table, as shown in SEQ ID NO.:1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com