Eimeria tenella gametophyte antigen gam59 gene and application thereof
A technology of Eimeria and antigen genes, applied in application, gene therapy, genetic engineering, etc., can solve the problem of no nucleotide sequence reporting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
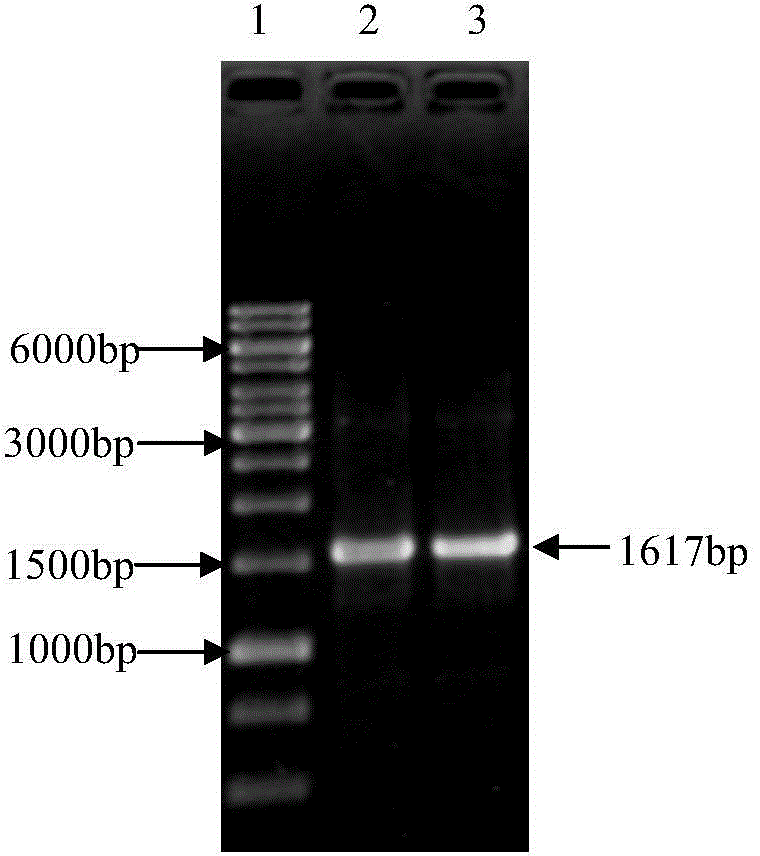
[0044] Example 1: Isolation of the Etgam59 gene from the Eimeria tenella gametophyte gene
[0045] The gametocytes of Eimeria tenella were isolated and purified by conventional methods, and the total RNA was extracted with an RNA extraction kit.
[0046] RT-PCR catch Etgam59 gene:
[0047] Referring to TaKaRa company (TaKaRa RNA LA PCR TM The method provided by Kit (AMV) Ver.1.1): the two-step RT-PCR kit amplifies the Etgam59 gene.
[0048] The Etgam59 gene was captured using an upstream primer (5'-CCCCGGGAACATGACTCGTCTCGCCGCC-3') and a downstream primer (5'-CGAATTCTTACTCAAATCCAAAAGAAGG-3').
[0049] The first step: reverse transcription reaction, the total system is 10μl: 0.5μl total RNA (≤500ng total RNA), 25mM MgCl 2 2μl, 10×RNA PCR Buffer 1μl, RNase Free dH 2 O 4.25 μl, dNTP 1 μl at a concentration of 10 mM, RNase Inhibitor 0.25 μl, 5 μ / μl AMV reverse transcriptase 0.5 μl and Oligo dT linker primer 0.5 μl. Reaction steps: reverse transcription at 42°C for 30 minutes; ...
Embodiment 2
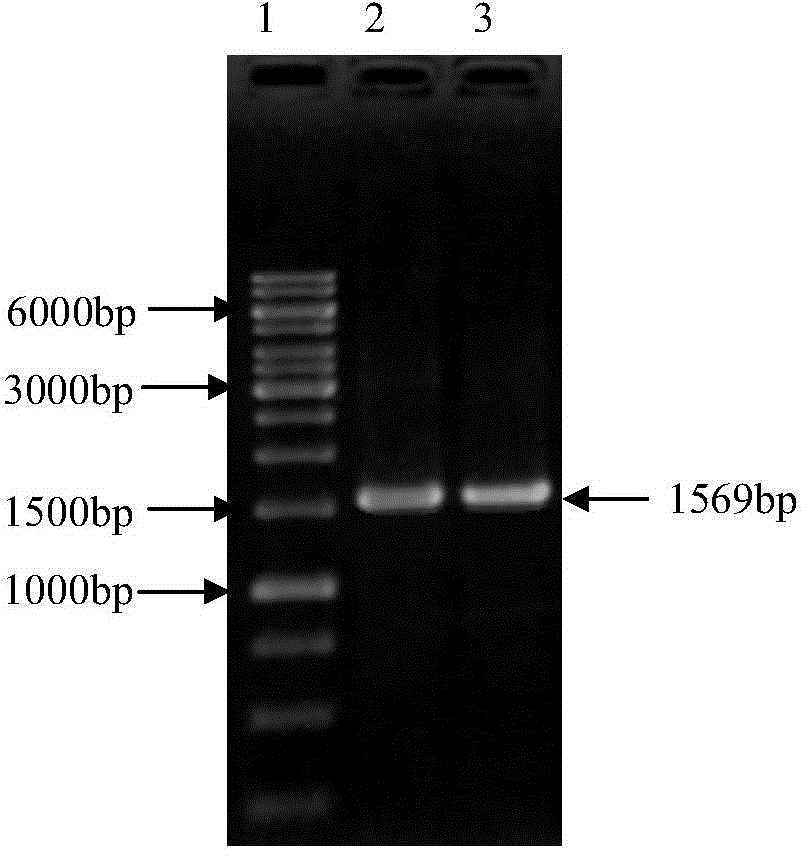
[0051] Embodiment 2: Amplification of expression fragment Etgam59
[0052] According to the ORF sequence of the Eimeria tenella Etgam59 gene and the restriction site of the plasmid pET28a (purchased from Invitrogen), after removing the signal peptide, specific primers were designed to amplify the gene fragment for expression.
[0053] According to the enzyme cutting sites on the plasmid pET28a, after sequence analysis of the Etgam59 gene, the signal peptide was eliminated, EcoRI and HindIII enzyme cutting sites were selected, and the specific primers were designed as: upstream primer (5'-CCGGAATTCATGCCCACGGCCATCCCC- 3') contains an EcoRI restriction site and 3 protective bases, and the downstream primer (5'-CCGAAGCTTTTACTCAAATCCAAAAGA-3') contains a HindⅢ restriction site and 3 protective bases.
[0054] The 50 μl PCR reaction system is as follows: 5 μl of 10× reaction buffer; 2 μl of upstream and downstream primers with a concentration of 10 μM, 8 μl of dNTP (2.5 mM); 30.5 μl...
Embodiment 3
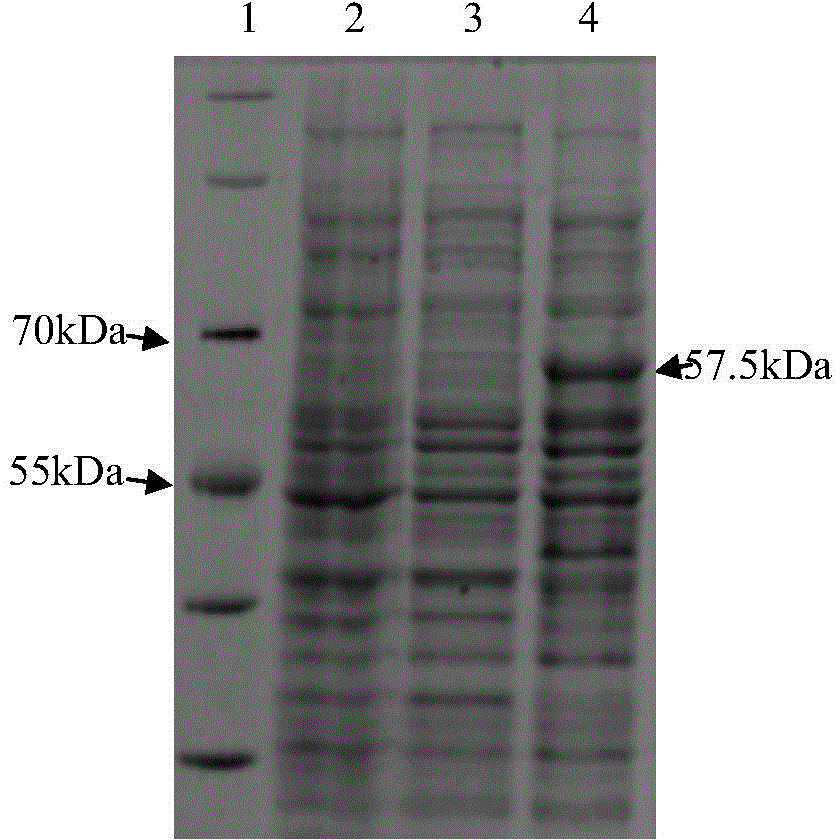
[0055] Embodiment 3: the construction of prokaryotic expression vector
[0056] Double digestion of pET28a:
[0057] EcoRI and HindⅢ are Fermentas products. 100 μl digestion system: 10 μl 10× FastDigest Green Buffer, 50 μl vector pET28a, 5 μl each of FastDigest EcoRI and FastDigest HindIII, 30 μl sterilized ddH 2 O. React at 37°C for 12 hours, perform 1.2% agarose gel electrophoresis, and recover the product from the gel.
[0058] The PCR product purified in double enzyme digestion embodiment 2:
[0059] EcoRI and HindⅢ are Fermentas products. 100 μl digestion system: 10 μl 10× FastDigest Green Buffer; 20 μl purified PCR (Etgam59) product; 5 μl each of FastDigest EcoRI and FastDigest HindIII; 60 μl sterilized ddH 2 O. React at 37°C for 12 hours, perform 1.2% agarose gel electrophoresis, and recover the digested product.
[0060] Link reaction:
[0061] Solution Ⅰ is a TaKaRa product. 9 μl double-digested vector pET28a, 1 μl double-digested PCR product, 10 μl solution ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com