PCR-RFLP method for detecting H-site mutation of PIK3CA gene
A PCR-RFLP, site mutation technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effects of good detection reproducibility and accuracy, improved accuracy, and improved sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
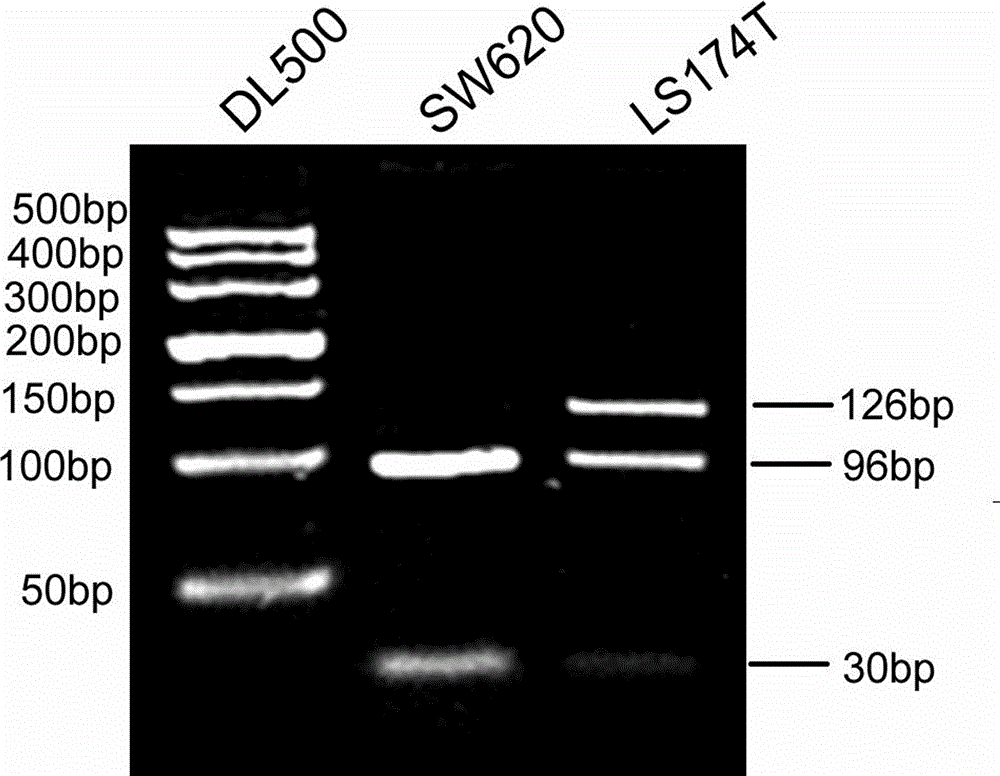
[0047] The PCR-RFLP method of the present invention detects the H1047R site mutation of the PIK3CA gene in colorectal cancer cells comprising the following steps:
[0048] (1) Genomic DNA extraction from samples
[0049] Human colorectal cancer cell lines SW620 and LS174T in the logarithmic growth phase were respectively taken, digested with trypsin, and counted. Take cells 1 x 10 6centrifuge at 1000 rpm for 5 min, discard the supernatant; extract the genomic DNA of the cells according to the operation steps of the Promega Whole Genome Extraction Kit; dissolve the extracted DNA in 20 μL ddH 2 O, measured at a wavelength of 260 nm and adjusted to a concentration of 0.5 μg / μL;
[0050] (2) PCR amplification of PIK3CA gene
[0051] Using about 100 ng of extracted genomic DNA as a template, PCR amplification was performed using the upstream primer 5'-GGAGTATTTCATGAAACAAATGAATGATGCG-3' and the downstream primer 5'-GAGCTTTCATTTTCTCAGTTATCTT-3' to obtain a 126bp amplified product;...
Embodiment 2
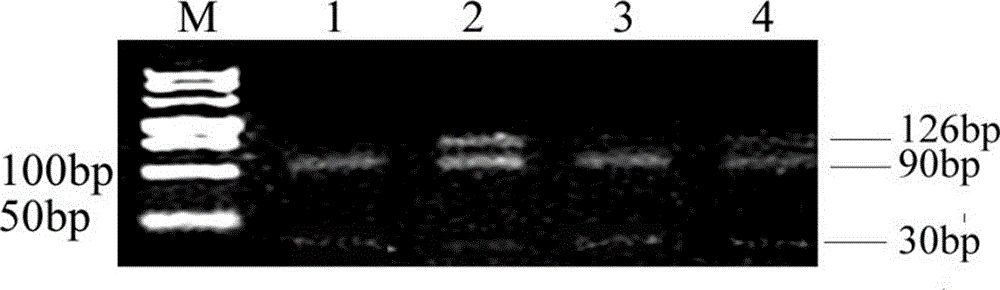
[0074] The PCR-RFLP method of the present invention detects the H1047R site mutation of the PIK3CA gene in the paraffin section specimen of the tumor tissue of a patient with colorectal cancer, comprising the following steps:
[0075] (1) Genomic DNA extraction from samples
[0076] Take paraffin sections (5 μm thickness) of tumor tissues from patients with colorectal cancer, extract the genomic DNA of the cells according to the operation steps of the OMEGA Paraffin Section DNA Extraction Kit, and finally elute the DNA with 40 μL of eluent for later use;
[0077] (2) PCR amplification of PIK3CA gene
[0078] Take 2 μL of the DNA eluate as a template, use the upstream primer 5'-GGAGTATTTCATGAAACAA ATGAATGATGCG-3' and the downstream primer 5'-GAGCTTTCATTTTCTCAGTTATCTT-3' to perform PCR amplification to obtain a 126bp amplification product;
[0079] The total volume of the PCR reaction is 25 μL, and the composition is:
[0080] wxya 2 O 16 μL
[0081] 10×buffer (with Mg 2+ )...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com