RAPD (random amplified polymorphic DNA) primers for differentiating pear varieties and application of primers
A variety and primer pair technology, applied in the field of molecular biology, can solve the problems of large operation volume, lack of practicability, and inability to visually display the primers of multiple varieties, and achieve the effect of simple operation and wide versatility.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Extraction of DNA.
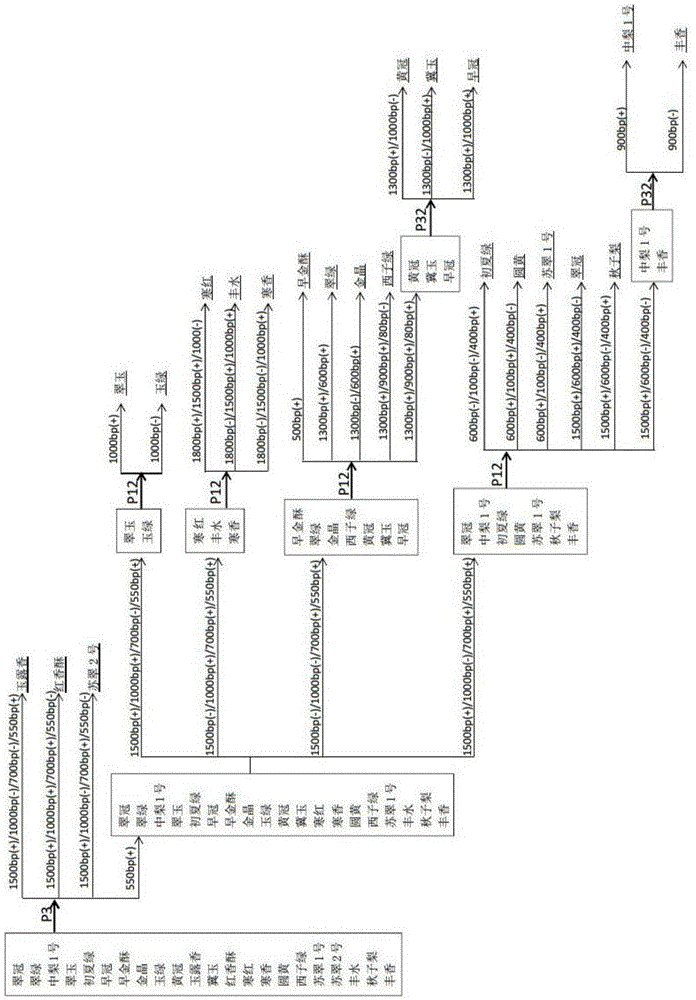
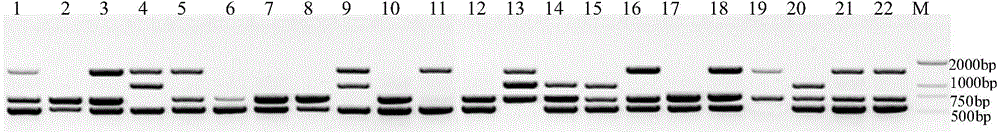
[0043] The pear variety samples of the present invention are all important pear varieties that are common in the market. These 22 varieties include: Cuiguan, Cuilu, Zhongli No. 1, Cuiyu, Chuxialv, Zaoguan, Zaojinsu, Jinjing, Yulu, Huangguan , Yuluxiang, Jiyu, Hongcrispy, Hanhong, Hanxiang, Yuanhuang, Xizigreen, Sucui No. 1, Sucui No. 2, Fengshui, Qiuzili, Fengxiang.
[0044] Take about 0.2g of young pear leaves as the material, grind with liquid nitrogen, add 500μl CTAB lysate (2% CTAB, 2M NaCl 2 , 20mM EDTA, 100mM Tris-HCl (PH=8.0) and 0.2% β-mercaptoethanol), transfer to a 1.5ml centrifuge tube, take out and cool in a 65°C water bath for 0.5-1h, add an equal volume of chloroform: isoamyl alcohol ( 24:1) After shaking the mixture gently, centrifuge the emulsion at 12000r / min for 8-10min, extract the supernatant with chloroform / isoamyl alcohol (24:1) for 1-2 times, add pre-cooled isopropanol 4 ℃ refrigerator for more than 3 hours, the c...
Embodiment 2
[0045] Example 2: RAPD-PCR reaction system and conditions.
[0046] RAPD-PCR reaction system is 20μl, containing 50ng genomic DNA, 2.0mmol L -1 MgCl 2 , 0.15mmol·L -1 dNTPs, 0.44 μmol L -1 10mer primers, 0.75U Taq DNA polymerase.
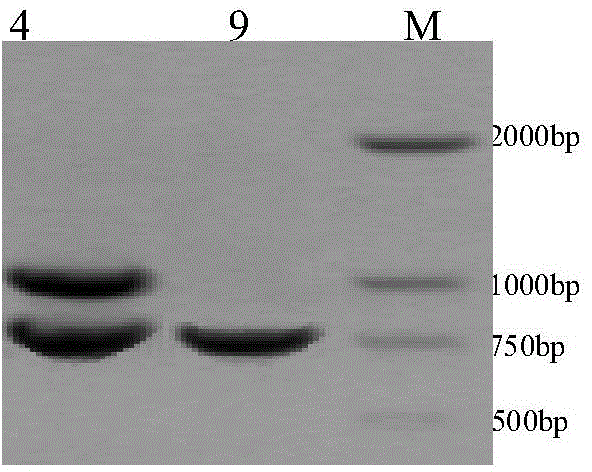
[0047] The RAPD-PCR amplification program was pre-denaturation at 94°C for 2min, denaturation at 94°C for 30S, annealing for 45S, extension at 72°C for 90S, 42 cycles, extension at 72°C for 10min, and storage at 4°C. Amplify on a PCR amplification instrument, and the amplified product adopts agarose gel electrophoresis (containing 1.2% ethidium bromide), 1 × TAE electrophoresis buffer (contains in 1L: 0.242g Tris, 0.057ml glacial acetic acid, 0.2 ml 0.25mol / L EDTA pH 8.0). Add 4 μl of 6× loading buffer (containing 0.25% bromophenol blue, 30% sucrose) to the amplified DNA sample, and perform electrophoresis at a constant voltage of 120V for 0.5-0.8 hours until the amplified DNA bands are clearly separated After that, the amplified bands were ob...
Embodiment 3
[0048] Example 3: Stringent screening of specific primers.
[0049] The three primers of the present invention are obtained through strict screening of 42 random primers originally designed. Firstly, 42 random primers were designed for the genome of the existing pear varieties, and 11 bases of RAPD random primers with good polymorphism, strong amplification and high stability were selected from them, and the band patterns were screened out by gradient PCR twice. Clear and reproducible primers. In detail, the gradient PCR reaction system is 20 μl, containing 50ng genomic DNA, 2.0mmol·L -1 MgCl 2 , 0.15mmol·L -1 dNTPs, 0.44 μmol L -1 10mer primers, 0.75U Taq DNA polymerase. The reaction was pre-denatured at 94°C for 2min, denatured at 94°C for 30S, annealed for 45S, extended at 72°C for 90S, 42 cycles, and extended at 72°C for 10min. According to 2 consecutive gradient PCRs, select the primers with clear bands and repeated PCR products, and the annealing temperature of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com