Rice seedling-stage salt-resistant gene qST2 and molecular marking method thereof
A technology of salt-tolerant genes and molecular markers, applied in the fields of rice stress resistance breeding and molecular genetics, to achieve the effect of facilitating cross-breeding and speeding up the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] The present invention will be further described below in conjunction with specific embodiment, wherein the method used is conventional method unless otherwise specified.
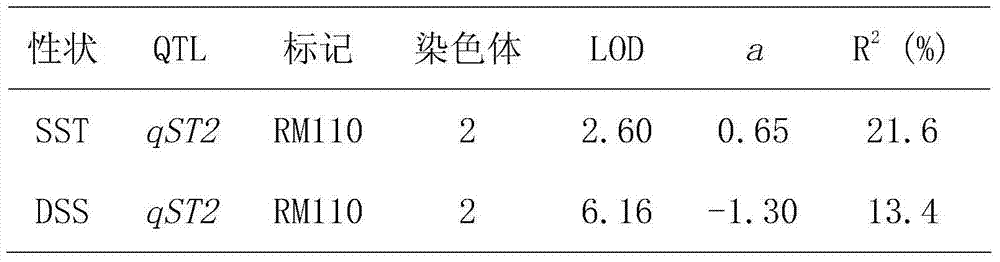
[0019] 1. Single-marker mapping of new salt-tolerant genes
[0020] 1. Test materials
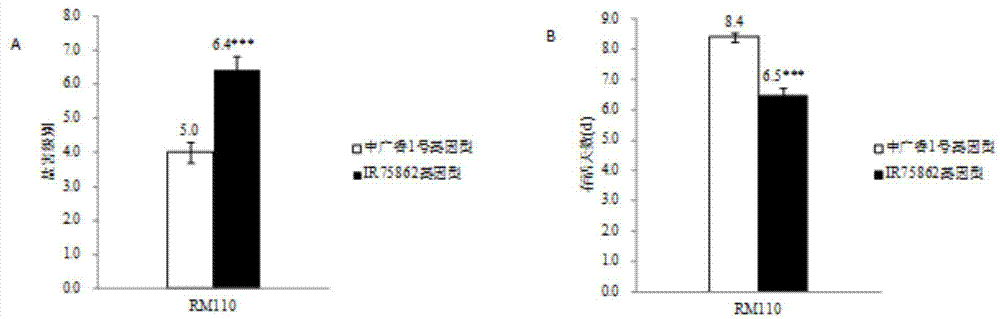
[0021] Using the glutinous rice variety IR75862 introduced from the International Rice Research Institute as the donor, and my country's high-yield and high-quality indica-type conventional rice Zhongguangxiang 1 as the recipient to cross and backcross, construct 200 BC 1 f 10 The backcross of the pedigree introduces the pedigree group.
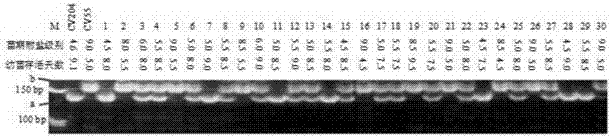
[0022] 2. DNA extraction, PCR amplification and gel electrophoresis
[0023] Referring to the DNA extraction method of Temnykh et al. (2000), genomic DNA was extracted from each individual plant. Polymerase chain reaction (PCR) was performed on the RM110 marker using the genomic DNA of each individual plant as a template. The products of the PCR reactions were separated by po...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com