Multifunctional oral vaccine based on chromosome recombineering
A chromosome and gene technology, applied in multivalent vaccines, genetic engineering, vaccines, etc., can solve problems such as anti-antibiotic resistance markers, plasmid instability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
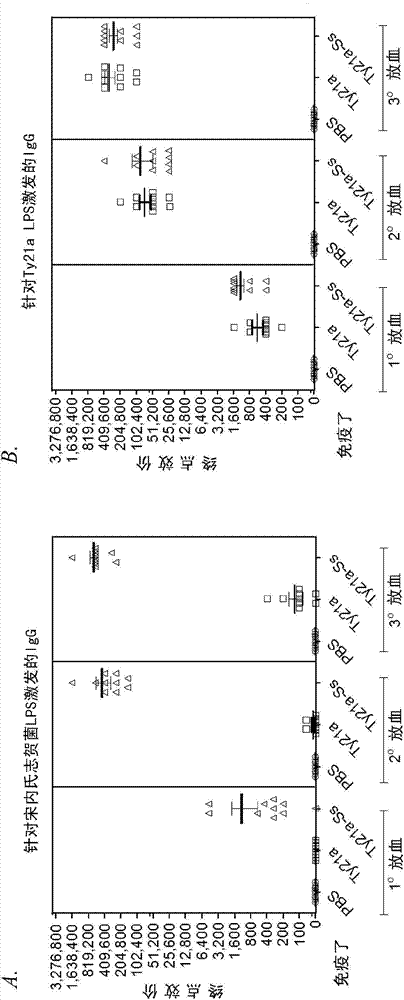
Image
Examples
Embodiment approach
[0077] In one embodiment, the invention includes a prokaryotic microorganism. In another embodiment, the prokaryotic microorganism is a bacterial species. Preferably, said prokaryotic microorganism is an attenuated strain of Salmonella. However, other prokaryotic microorganisms such as attenuated Escherichia coli, Shigella, Yersinia, Lactobacillus, Mycobacteria, Listeria or Vibrio Strains are also encompassed by the invention. Exemplary suitable microbial strains include, but are not limited to, Salmonella typhimurium, Salmonella typhimurium, Salmonella dublin, Salmonella enteritidis, Escherichia coli, Shigella flexneri, Shigella sonnei , Vibrio cholerae (Yamamoto, et al. (2009) Gene 438:57-64), Pseudomonas aeruginosa (Lesic, et al. (2008) BMC Mol Biol 9:20), Yersinia pestis Yersinia pestis (Sun, et al. (2008) Applied and Env Microbiol 74:4241-4245) and Mycobacterium bovis (BCG). Notably, λred does not function in mycobacteria, but another phage (e.g. Che9c gp61) has been us...
Embodiment 1
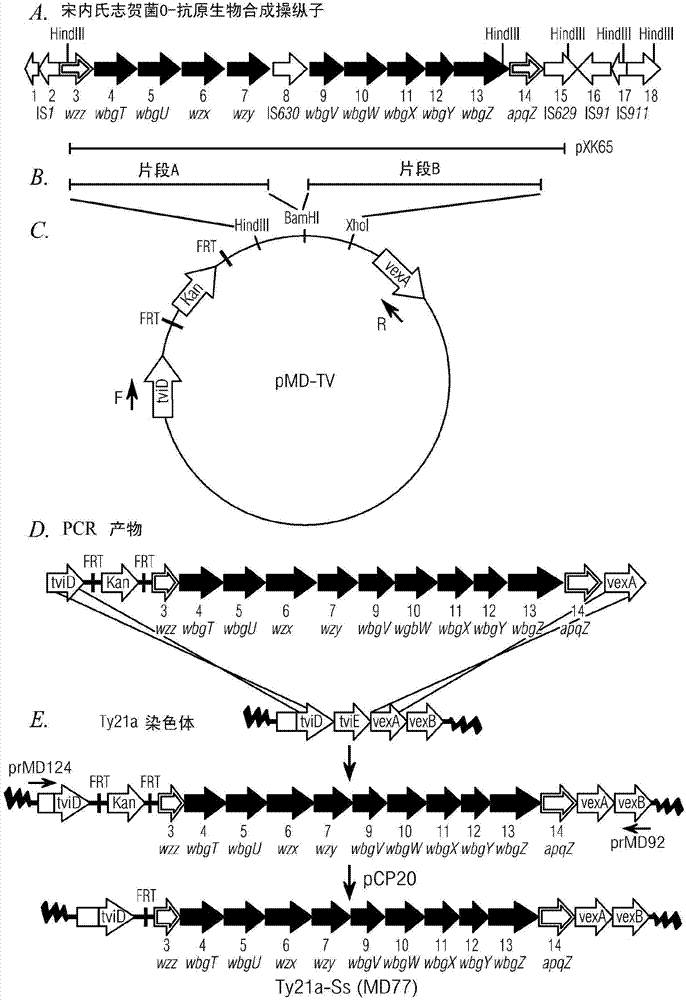
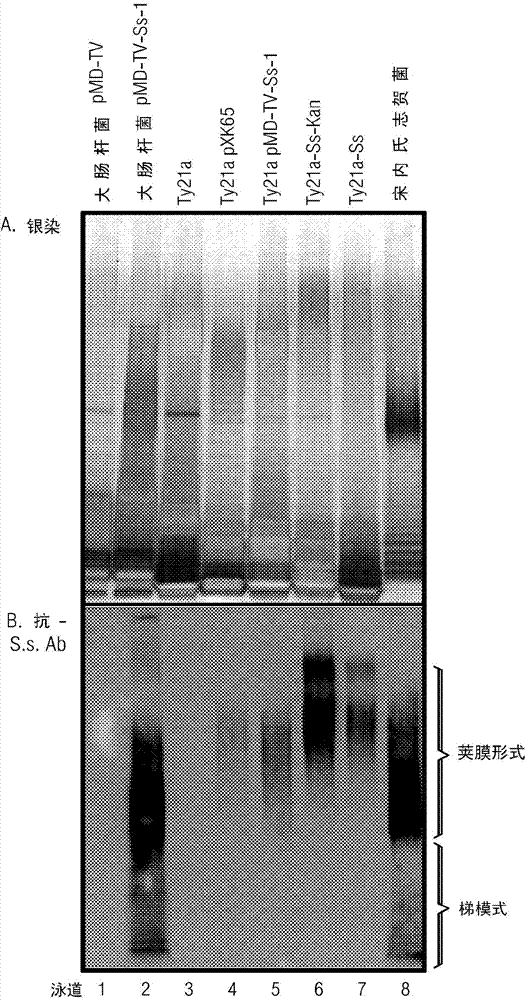
[0170] Example 1: Cloning of minimal essential Shigella sonnei O-antigen biosynthesis genes
[0171] A large 30 kb region and flanking sequences encoding the biosynthesis of the S. sonnei type I O-antigen were pre-identified (initially by deletion analysis) to determine the minimal essential sequence for O-antigen expression. Together with DNA sequence analysis of this region, these deletion data indicated the presence of a putative promoter and 10 orf( figure 1 A contiguous 12.3 kb region in orf 4-13) is required for O-antigen biosynthesis in Shigella sonnei. These deletion studies also suggest that wzz (ofr3), which is typically involved in the regulation of O-antigen chain length, is not required for type 1 expression in Ty21a, but the latter part of wzz apparently contains the putative type 1 O-antigen biosynthesis operon promoter ( Xu, et al. (2002) Infect and Immun 70:4414-4423.
[0172] Construction of pMD-TV plasmid
[0173] Cloning is performed using standard mol...
Embodiment 2
[0182] Example 2: Integration of linear Shigella DNA into the Ty21a chromosome
[0183] Datsenko and Wanner (Datsenko et al. (2000) Proc. Natl. Acad. Sci. USA, 97:6640-6645) previously described a method based on the highly efficient lambda phage red recombination system, which enables Antibiotic resistance gene recombination replaces E. coli chromosomal sequences to generate specifically targeted gene mutations, generated by PCR using primers containing a 36-50 bp extension homologous to the targeted mutated gene. The λred system includes β, γ and exo genes, the products of which are called Beta, Gam and Exo, respectively (Murphy (1998) J. Bacteriol., 180:2063-2071). Gam inhibits the host RecB, C, D exonuclease and SbcC, D nuclease activities, resulting in exogenously added linear DNA is not degraded. The Exo protein is a dsDNA-dependent exonuclease that binds the ends of each strand while degrading the other strand in the 5' to 3' direction. Beta binds the resulting ssDN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com