Kit and primer pair group for detecting drug-induced deafness susceptibility gene mutation site
A technology for drug-induced deafness and mutation sites, applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of false positive sensitivity, cumbersome detection process, cross-contamination, etc. Efficiency, low price, low instrument requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Detection of Two Mutation Sites in Human Mitochondrial Genome 12S rRNA Gene
[0049] 1. Primer design
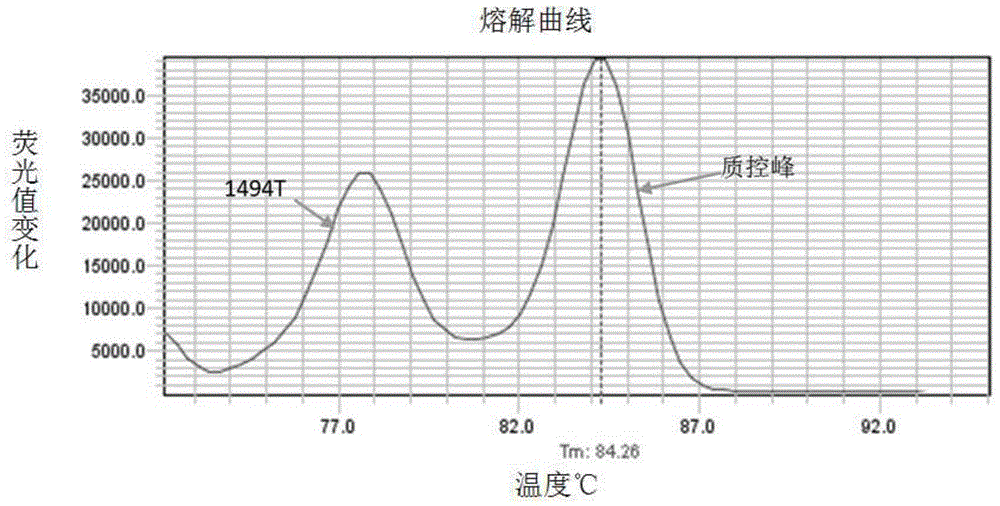
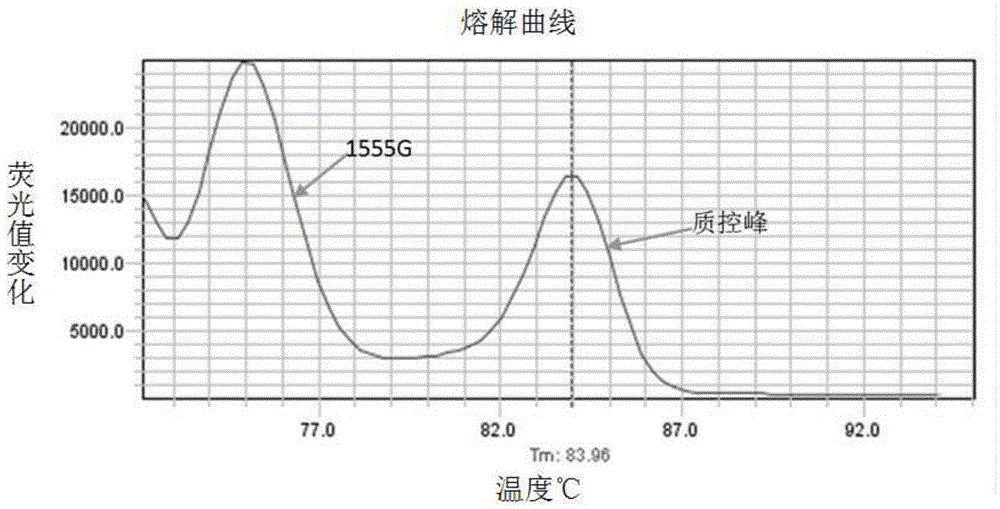
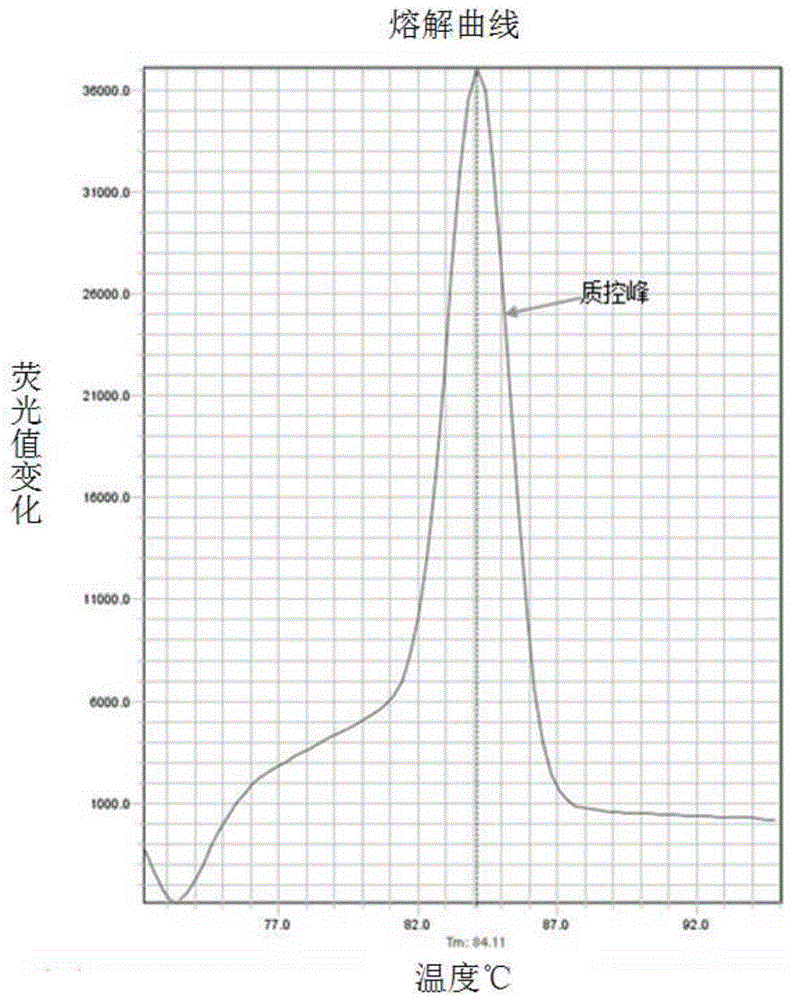
[0050] According to the principle of site-specific primer design, the mutation site was set as the 3' end of the primer, and the mutation site mutation was introduced near the 3' end to improve specificity. The primer 5.0 software was used to design the C1494T and A1555G mutation sites respectively. primers. The design of the primers needs to ensure that only the mutant template can be amplified in a certain amplification system, but not the normal template; and ensure that the Tm values of the PCR products of the C1494T and A1555G mutant templates can be separated during melting curve analysis (such as Tm The difference is above 2°C); while avoiding non-specific Tm peaks caused by by-products.
[0051] Taking the C1494T and A1555G mutations in mitochondrial 12S rRNA as an example, according to the human mitochondrial genome nucleic acid sequence (NC_012...
Embodiment 2
[0080] Example 2 Detection of C1494T and A1555G mutations in clinical samples using primer pairs of the present invention
[0081] Using the primer pair set of Example 1 of the present invention, 200 samples from the general population were detected, and the C1494T and A1555G mutations in the samples were detected. All samples were sequenced and verified by the gold standard sequencing method. 2 cases of positive samples C1494CT, 4 cases of positive samples A1555AG, and 194 cases of negative samples were detected. The kit of the present invention detected 200 cases of clinical samples compared with the gold standard sequencing results. The detection results were consistent, 0 cases were missed, and the accuracy was 100%; For other genotype positive and negative samples that are not within the detection range of this kit, the detection results of this kit are all negative, with a sensitivity of 100% and a specificity of 100%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com