Protein subcellular localization and prediction method realized by using nearest-neighbor retrieval
A technology of subcellular localization and prediction method, which is applied in the field of protein subcellular localization prediction realized by nearest neighbor retrieval, to achieve the effects of strong model adaptability, effective acquisition, and high overall accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0043] The present invention will be further described below in conjunction with the accompanying drawings and embodiments.
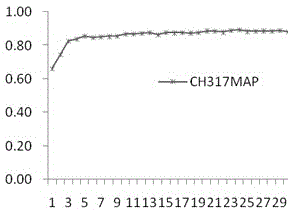
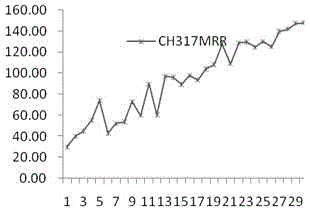
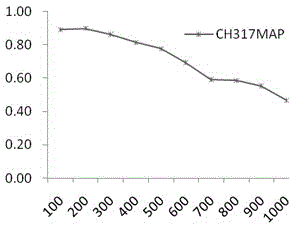
[0044] 1 Selection of test data set
[0045]Take the dataset containing 317 apoptotic protein sequences obtained from the SWISS-PROT database as an example. 317 protein sequences, distributed in 6 intervals, including 112 cytoplasmic proteins, 55 membrane proteins, 34 mitochondrial proteins, 17 secreted proteins, and 52 nuclear proteins Strips, endoplasmic reticulum proteins (Endoplasmicreticulumproteins) 47.
[0046] 2 Experimental evaluation methods and indicators
[0047] There are three common predictive evaluation methods: Resubstitution, K-fold cross validation and Jackknife. For the self-compatibility test, the test set contains the sequence to be predicted, and it can be predicted that the detection success rate of the method in this paper is 100%. Compared with the K-fold cross-test, the knife-cut test uses a one-to-many prediction model, w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com