Small GTP binding protein gene TaRab18 and expression vector and application thereof
A technology combining proteins and expression vectors, applied in the field of genetic engineering, can solve the problems of variety resistance loss, single large-scale planting, etc., and achieve the effect of improving resistance and stripe rust resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1 is subjected to the cloning of the gene TaRab18 with GTP / GDP binding effect induced by stripe rust
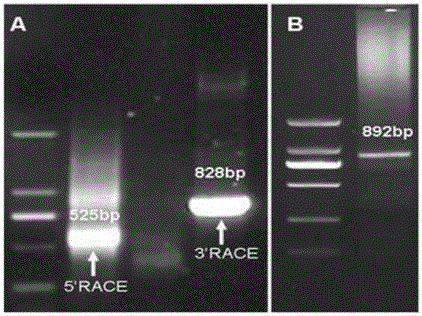
[0027] In order to clone the resistance-related genes in the Yr26 disease resistance pathway, gene microarray hybridization was used to screen the differentially expressed genes in stripe rust-resistant wheat 92R137 and stripe rust-susceptible wheat Yangmai 158. An EST probe Ta.1763.2.S1_at with 10-fold up-regulated expression was obtained from multiple screenings. Based on this probe sequence, the amplification product of expected length was obtained by RACE ( figure 1 A), the sequence length after splicing is 1015bp. Full-length primers P1 (CCAGCCATGGACTCTTCTTC, SEQ ID NO.3) and P2 (AGCTGAAAGCTGCCAAGGTA, SEQ ID NO.4) were designed according to the spliced sequence. Using the disease-resistant wheat 92R137cDNA as a template, an amplified product with an expected length of 826bp was obtained by RT-PCR ( figure 1 B), and it is recovered, cloned, and sequ...
Embodiment 2
[0028] Embodiment 2 TaRab18 gene is subjected to the expression characteristic analysis of stripe rust induction
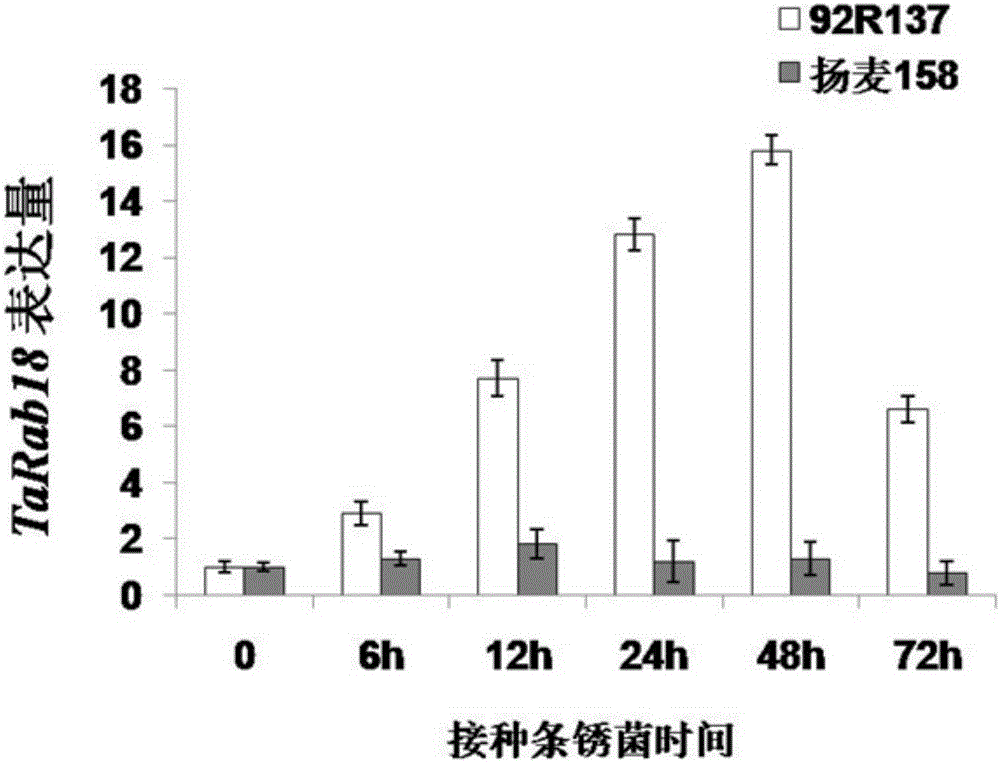
[0029] In order to study the expression pattern of TaRab18 in stripe rust-resistant materials, the RNA reverse transcription cDNA of the resistant material 92R137 and the susceptible material Yangmai 158 induced by stripe rust for 0, 6, 12, 24, and 72 hours were used as templates. Real-time fluorescent quantitative PCR (Q-PCR) analysis was performed using P3 (CGCAGCCGGACTTCGACTACC, SEQ ID NO.5) and P4 (CCCAGACGGCGAGCTTGAGC, SEQ ID NO.6) as primers. The PCR program is as follows: the PCR reaction is amplified on a real-time fluorescent quantitative PCR instrument (MyIQ, Bio-Rad Company, USA) and the fluorescence is detected. The 20uLPCR reaction system contains 10uL of 2×SYBRGreenPCRMasterMix, 0.5μM primers P3 and P4, and 2uL of reverse transcription cDNA template. The amplification parameters were: 95°C for 10 minutes, then 95°C for 15 seconds, 60°C for 30 second...
Embodiment 3
[0030] Example 3 Construction of TaRab18 overexpression vector and its transformation into common wheat Yangmai 158 and identification of resistance to stripe rust
[0031]Using the cDNA from 92R137 as a template, the primers P5 (ATCCCGGGTATGGTTGAGTGCACAATGG, SEQ ID NO.7) and P6 (ATAGGTACCGAGCCTAGATCTTCAGCAGA, SEQ ID NO.8) across the ORF were designed with the full-length sequence of the TaRab18 gene, and P5 has a restriction site for BamH1, P6 has a restriction site for KpnI. PCR amplification was performed using primer pairs P5 and P6, and amplified fragments were recovered. The amplified product was double digested with BamH1 and KpnI, and the digested product was inserted into the vector pBI220 (JeffersonRA, KavanaghTA, BevanMW.GUSfusions:beta-glucuronidaseassensitiveandversatilegenefusionmarkerinhigherplants.EMBOJ.1987,6:3901- 3907.), put TaRab18 at the multiple cloning site behind the 35S promoter, and replace the GUS gene contained in the vector itself. Thus, the targ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com