PSR detecting method for pseudomonas aeruginosa, special primers and kit
A Pseudomonas aeruginosa and kit technology, applied in the biological field, can solve problems such as the advent of PSR-specific primers and kits for Pseudomonas aeruginosa, and achieve the goal of being suitable for wide-scale popularization and application, broad market prospects, and simple operation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1. Primer design for PSR detection of Pseudomonas aeruginosa
[0047] Obtain the Pseudomonas aeruginosa sequence (GenBank number: NC_002516.2) from the U.S. gene database retrieval, carry out homology analysis by BLAST software, obtain the specific conserved target sequence ToxA gene of Pseudomonas aeruginosa (sequence 1 in the sequence table ), and then use the software PrimerdesignV4 to design primers for PSR detection of Pseudomonas aeruginosa according to the conserved target DNA sequence.
[0048] Table 1 is used for the primer that Pseudomonas aeruginosa is carried out PSR detection
[0049] Primer name
[0050] The above sequence 2-sequence 5 were artificially synthesized.
Embodiment 2
[0051] Embodiment 2, establishment of the PSR detection method of Pseudomonas aeruginosa of the present invention
[0052] Carry out PSR detection to Pseudomonas aeruginosa with the five primers that are used for the PSR detection of Pseudomonas aeruginosa obtained in Example 1, to obtain the best reaction system and reaction conditions, the specific methods are as follows:
[0053] 1. Determination of the optimal reaction primer concentration
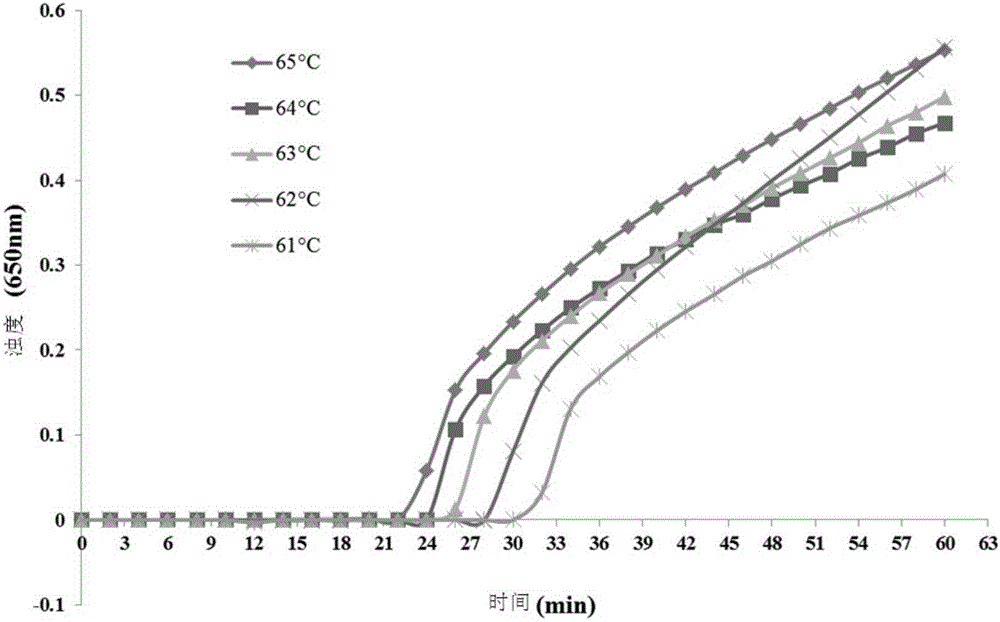
[0054] Under the same reaction conditions (keep at 63°C for 60 minutes), add different concentrations of primer combinations to the reaction system to determine the optimal reaction primers and their concentrations, as follows:
[0055] 1. PSR response
[0056] Template preparation: Promega's WizardGenomicDNApurificationKit commercial DNA extraction kit was used to extract the genomic DNA of Pseudomonas aeruginosa;
[0057] PSR reaction system: with Pseudomonas aeruginosa genomic DNA as a template, PSR amplification is carried out un...
Embodiment 3
[0079] Embodiment 3, the specificity and sensitivity detection of the PSR detection method of Pseudomonas aeruginosa of the present invention
[0080] One, the specificity detection of the PSR detection method of Pseudomonas aeruginosa of the present invention
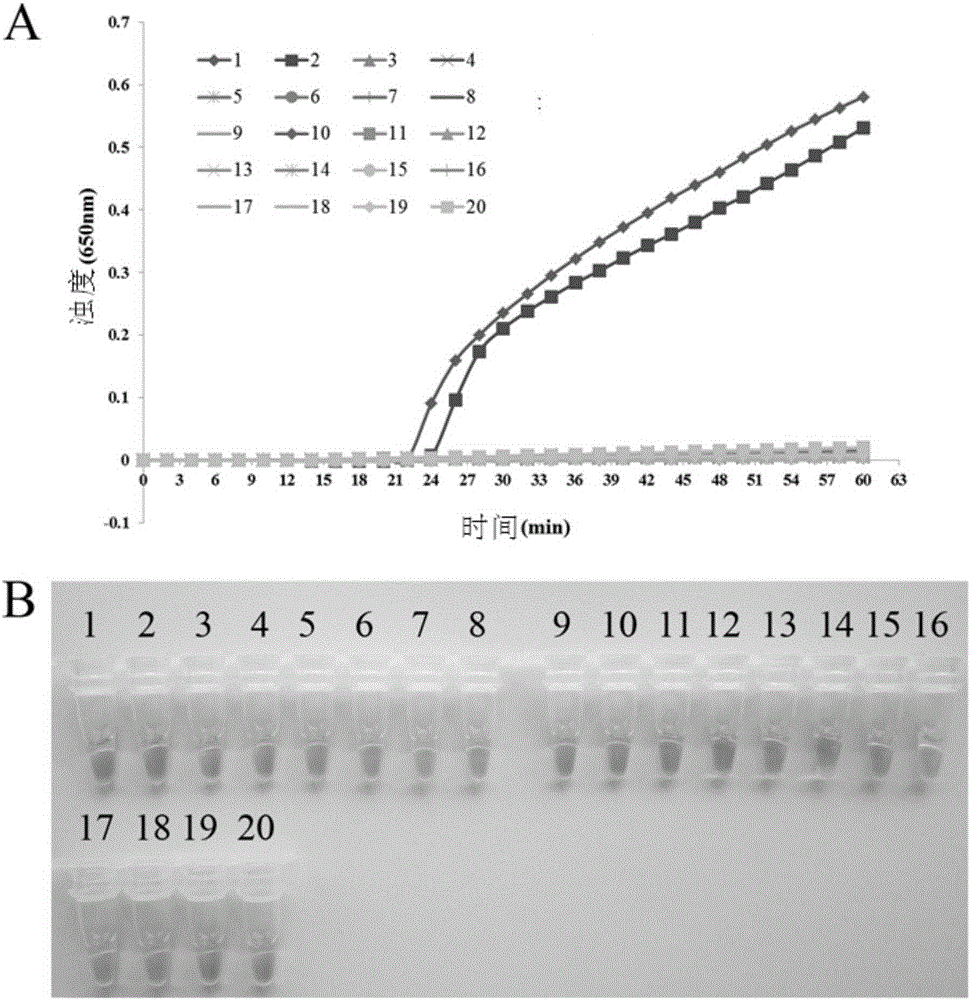
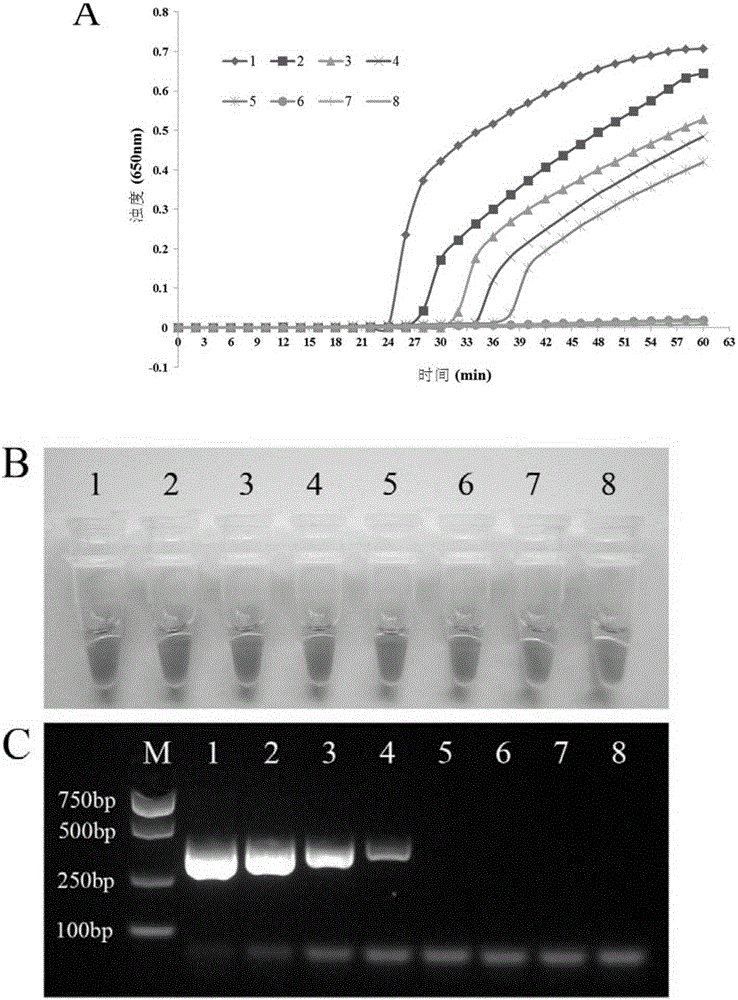
[0081] 1. PSR response
[0082] 模板制备:提取如下菌株的基因组DNA:1,P.aeruginosaATCC15442;2,P.aeruginosaCMCC10539;3,PseudomonasfluorescensCGMCC1.1802;4,Burkholderiapseudomallei029;5,PseudomonasgeniculateCGMCC1.871;6,PseudomonasmendocinaCGMCC1.593;7,Pseudomonasputida2309;8, KlebsiellapneumoniaeATCC2146;9,Streptococcuspneumoniae112-07;10,Mycobacteriumtuberculosis005;11,Staphylococcusaureus2740;12,Acinetobacterbaumannii12101;13,Escherichiacoli44825;14,Shigellaflexneri4536;15,StenotrophomonasmaltophiliaK279a;16,Legionellapneumophila9135;17,HaemophilusinfluenzaATCC49247;18,Salmonellatyphi9275;19,ProteusvulgarisCMCC49027;
[0083] PSR reaction system: the best detection system determined with embodiment 2 is the same;
[0084] The PSR re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com