System and application for rapid analysis of RNA functional elements in vivo
A functional element and rapid analysis technology, applied in the field of genetic engineering, can solve problems such as the inability to quickly find mRNA cis-acting elements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
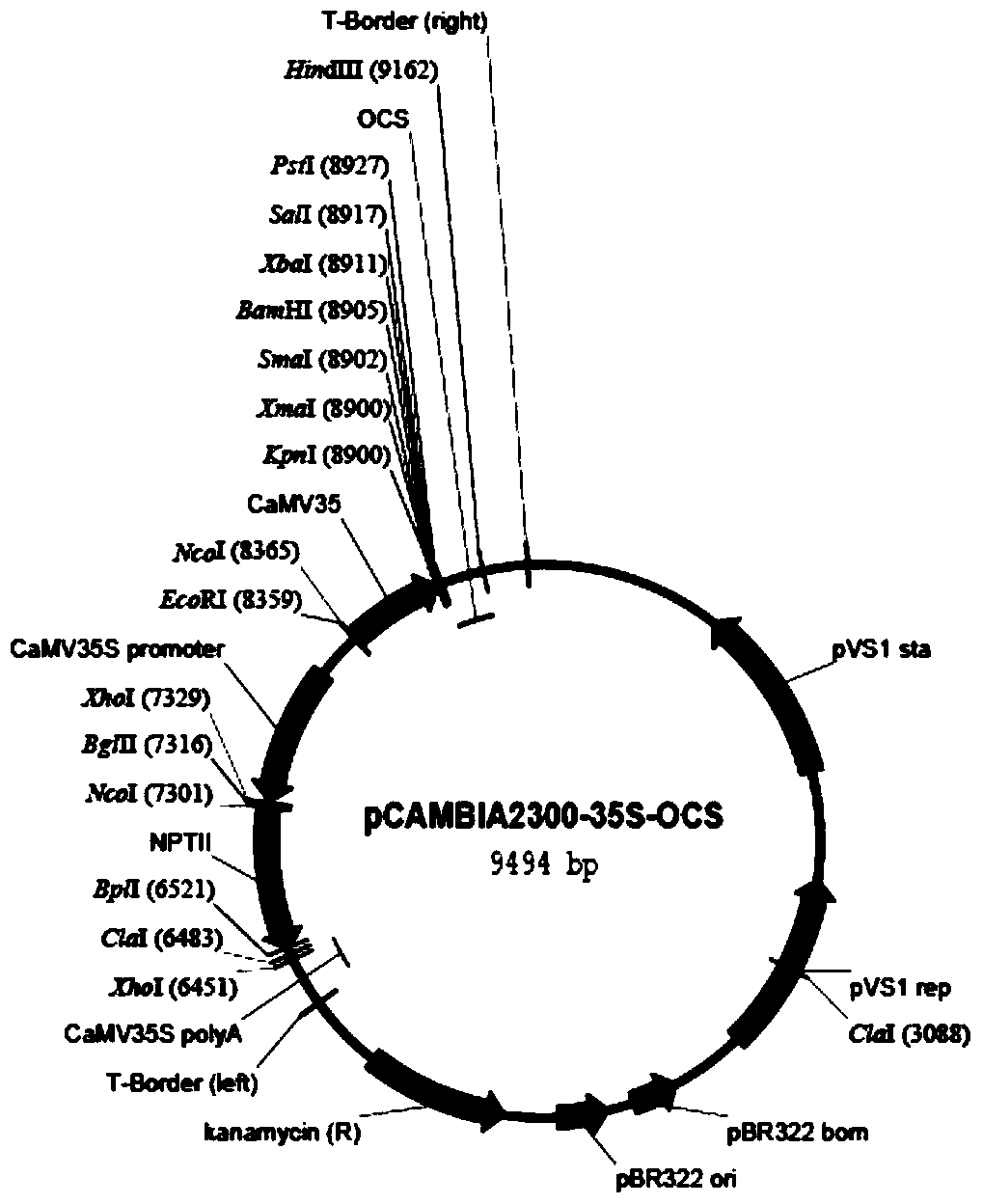
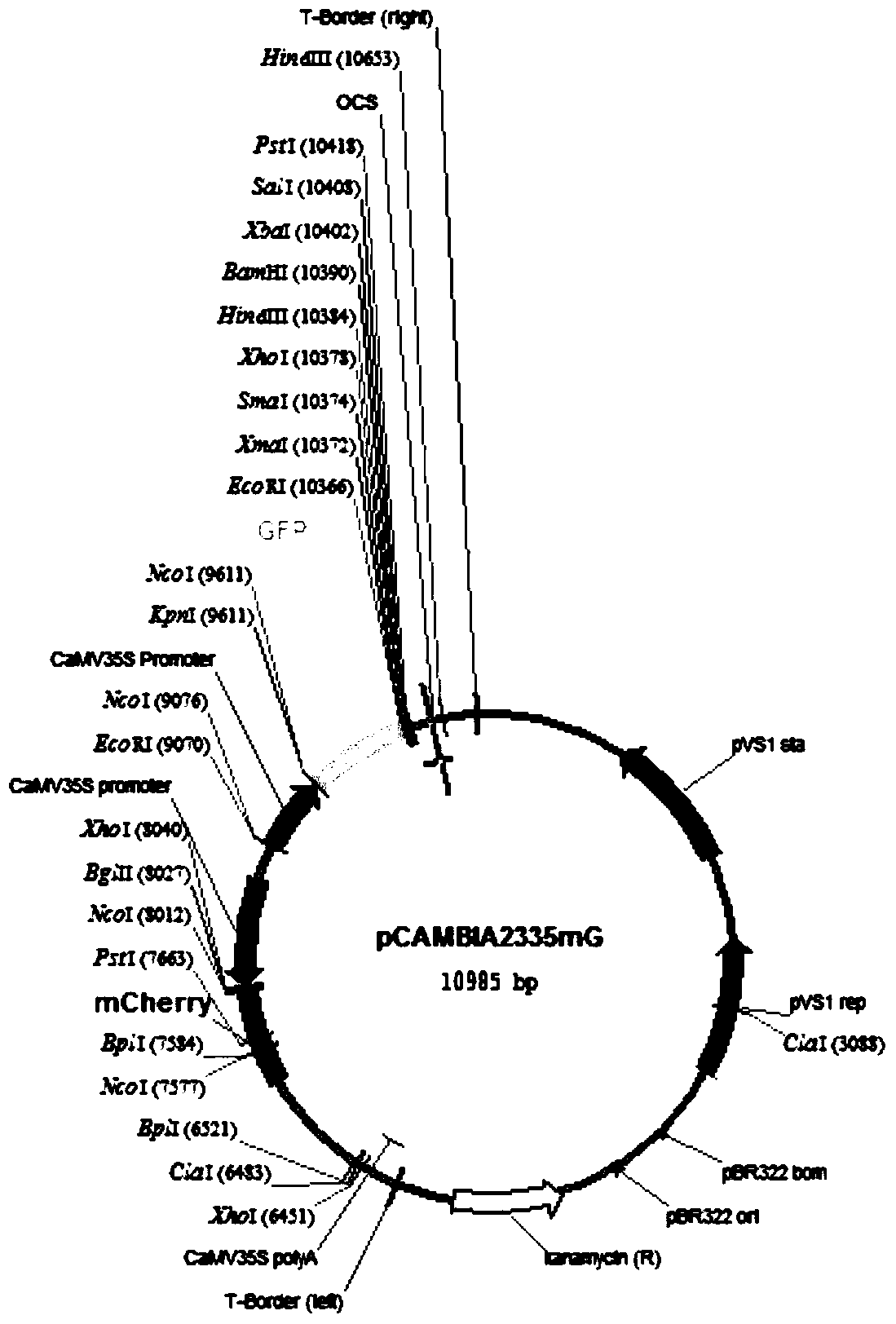
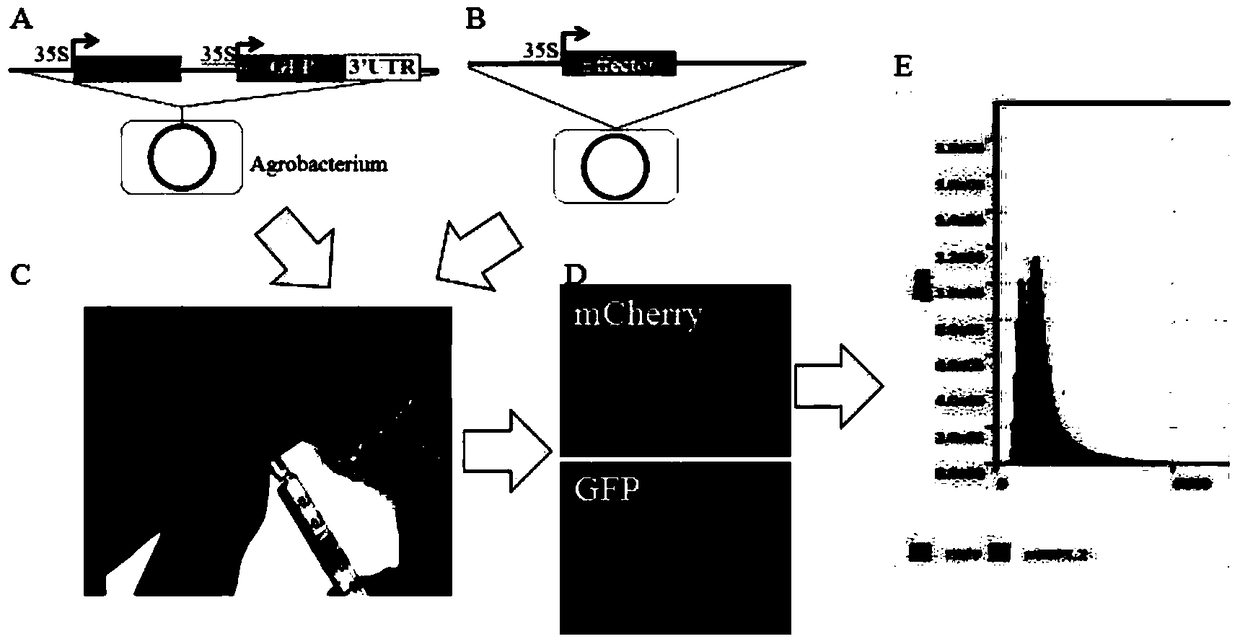
[0030] This embodiment provides a system for rapid analysis of RNA functional elements in vivo, for figure 1 The pCAMBIA2300-35S-OCS vector shown is transformed, and its resistance gene NPTII is excised with the restriction endonuclease XhoI, and connected into the mCherry fluorescent protein gene; in the multi-cloning site, there are two restriction sites of KpnI and XbaI The GFP fluorescent protein gene is interlinked, so that the two fluorescent proteins are driven by two different 35S strong promoters on the same carrier, forming a dual fluorescent reporter carrier, named pCAMBIA2335mG, referred to as p35mG, such as figure 2 shown.
[0031] The specific method of constructing the vector is as follows:
[0032] 1. Primers
[0033] Amplify mCherry:
[0034] forward primer
[0035] tctctcgagctttcgcagatctgtcgatcgaccatggtgagcaagggcgaggaggataacatggccatcatcaaggagttc
[0036] reverse primer agcctcgagtcacttgtacagctcgtccatgccgccggtggag
[0037] Amplified GFP:
[0038] Forwar...
Embodiment 2
[0087] This example provides a system for rapid analysis of RNA functional elements in vivo, which can accurately identify fragments that respond to ethylene signals and mediate translation inhibition.
[0088] The classic ethylene response is the well-known triple reaction, which is manifested in that when ethylene is applied, the etiolated seedlings show shorter and thicker hypocotyls, shorter roots, and intensified apical hooks. Previous studies found that the transgenic plants (G1U) overexpressing the full-length 3'UTR of EBF1 grown for 3 days on the medium containing 10 μM ACC (a precursor of ethylene biosynthesis) had obvious ethylene insensitivity. type, its hypocotyl length and root length are significantly longer than the wild type, and the top hook disappears, which has been disclosed in Chinese Patent No. ZL 201110110101.0.
[0089] Similarly, when we overexpress the above-mentioned fragments proved to have translation inhibition function by the detection system of ...
Embodiment 3
[0094]This embodiment provides a system for rapid analysis of RNA functional elements in vivo, which can detect the inhibitory effect of effector proteins on RNA translation.
[0095] The system provided in this example for rapid analysis of RNA functional elements in vivo can detect fragments that respond to ethylene signals and mediate translational repression, and the conclusions drawn are consistent with the phenotype of overexpressed 3'UTR fragments in Arabidopsis thaliana. Therefore, this system can be widely used in rapid analysis of RNA functional elements in plants, and can detect the regulation of different signals on 3'UTR.
[0096] In the tobacco transient expression system of this example, multiple Agrobacteria containing different plasmids can be transformed simultaneously, so this system can also be used to study the regulation of effector proteins on RNA. EIN5 (Ethylele Insensitive5, Potuschak et al., Plant Cell, 2006; Olmedo et al., PNAS, 2006) is known as a P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com