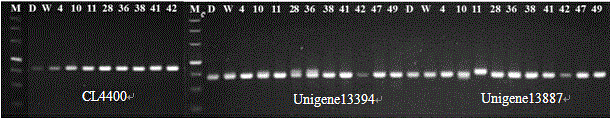
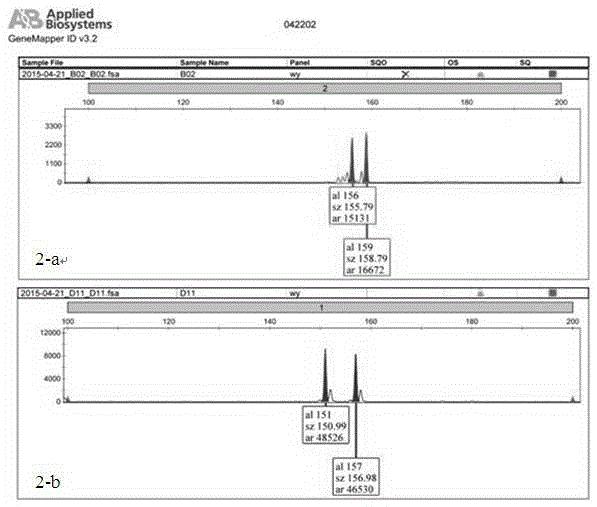
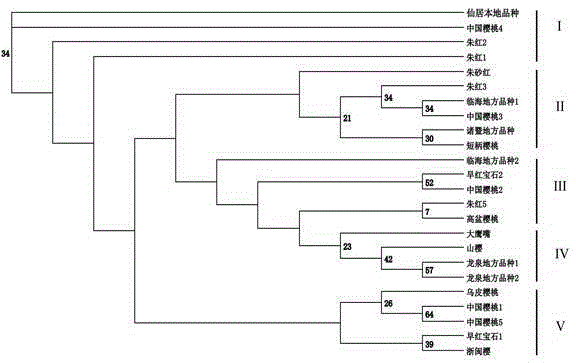
Primers for detecting SSR (simple sequence repeat) marks of prunus pseudocerasus
A technology for detecting primers and cherries, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve the problem of insufficient number of molecular markers in cherries
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] In the present invention, the specific method of using the SSR marker developed by the transcriptome of Chinese cherry "Short handle cherry" is:
[0057] 1. DNA extraction
[0058] (1) Preparation of DNA extraction solution: 2% CTAB, 0.1 MTris, 20 mM EDTA, 1.4 M NaCl, pH 8.0; DNA dissolution buffer (ie TE buffer): 10 mM Tris; 1 mM EDTA; pH=8.0.
[0059] (2) Chinese cherry material is processed as follows:
[0060] ① Add 4 mL of DNA extraction solution and 80 μL of β-mercaptoethanol to a 10 mL centrifuge tube, and place it in a 65°C water bath to preheat for 10 minutes;
[0061] ②Take 1g of fresh leaves, put a little PVP in the mortar, add liquid nitrogen to grind thoroughly; transfer the ground leaves to a centrifuge tube containing the extract, mix them well and put them in a water bath at 65°C for 30 minutes. Shake once every 10 minutes to fully lyse the leaf cells;
[0062] ③After the water bath, take out the centrifuge tube and cool it to room temperature; add an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com