Detection method of chromosome copy number variation
A technology of chromosome copy number and detection method, which is applied in the field of detection of chromosome copy number variation, can solve the problems that cannot be ruled out, high false positive results, etc., and achieve the effects of avoiding irreversible losses, simple operation, and avoiding false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1c
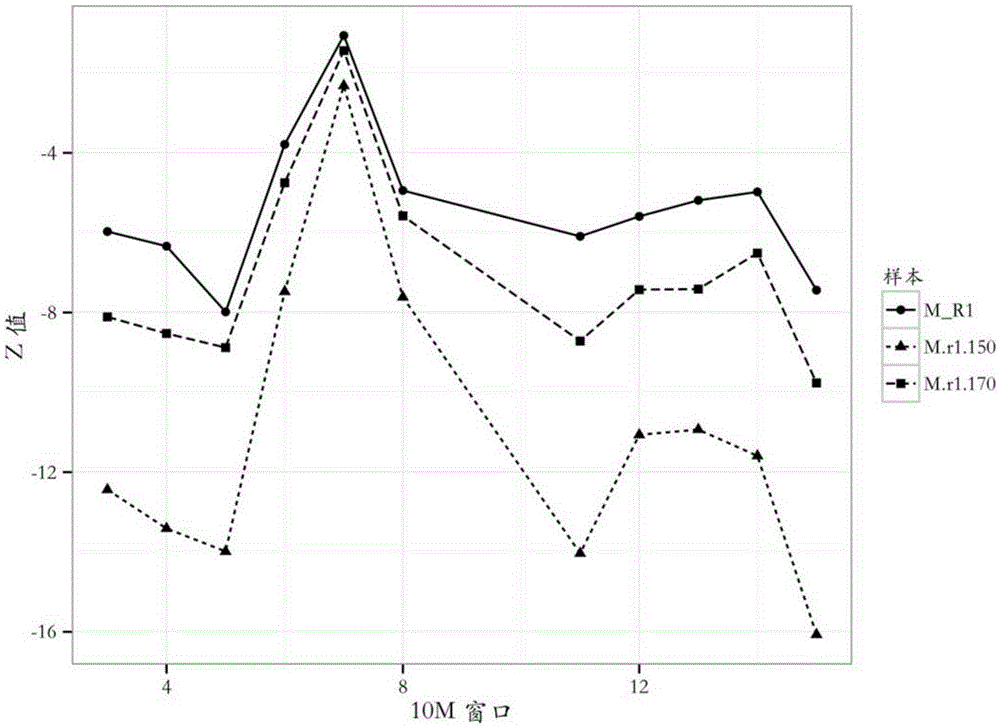
[0039] The judgment of embodiment 1cnv source fetus
[0040] A male fetus (XY) is used to simulate fetal Turner syndrome (XO), the absence of an X chromosome.
[0041] (1) Take 5mL of venous blood from 10 pregnant women with male fetuses, centrifuge at 1000rpm / min for 10min, take out the upper layer of plasma and then centrifuge at 8000rpm / min for 10min, absorb the upper layer of plasma, and use DNA from Qiagen Extraction kit QIAampDNABloodMiniKit (cat.no.51104) carries out DNA extraction. Obtain the free DNA fragment extracted from plasma, write down the mixed sample of this group of samples as M, as the sample to be tested;
[0042] (2) Take the same method as step (1) to extract cell-free DNA from 15 normal females as a control sample.
[0043] (3) All samples were subjected to high-throughput sequencing (see Chinese patent application CN201510039737.9) to obtain DNA sequence information.
[0044] (4) Align the DNA sequence of each sample to the human reference genome seq...
Embodiment 2c
[0053] The judgment of embodiment 2cnv source parent
[0054] Males (XY) were used to model Turner syndrome of maternal origin (XO).
[0055] (1) According to the method of step (1) of Example 1, the plasma cell-free DNA of a normal male is extracted, and the sample is M1, which is used as the sample to be tested.
[0056] (2) This step is the same as step (2) in Example 1.
[0057] (3) This step is the same as step (3) in Example 1.
[0058] (4) This step is the same as step (4) in Example 1.
[0059] (5) This step is the same as step (5) in Example 1, but wherein, N=166.
[0060] (6) This step is the same as step (6) in Example 1, but wherein, N=166.
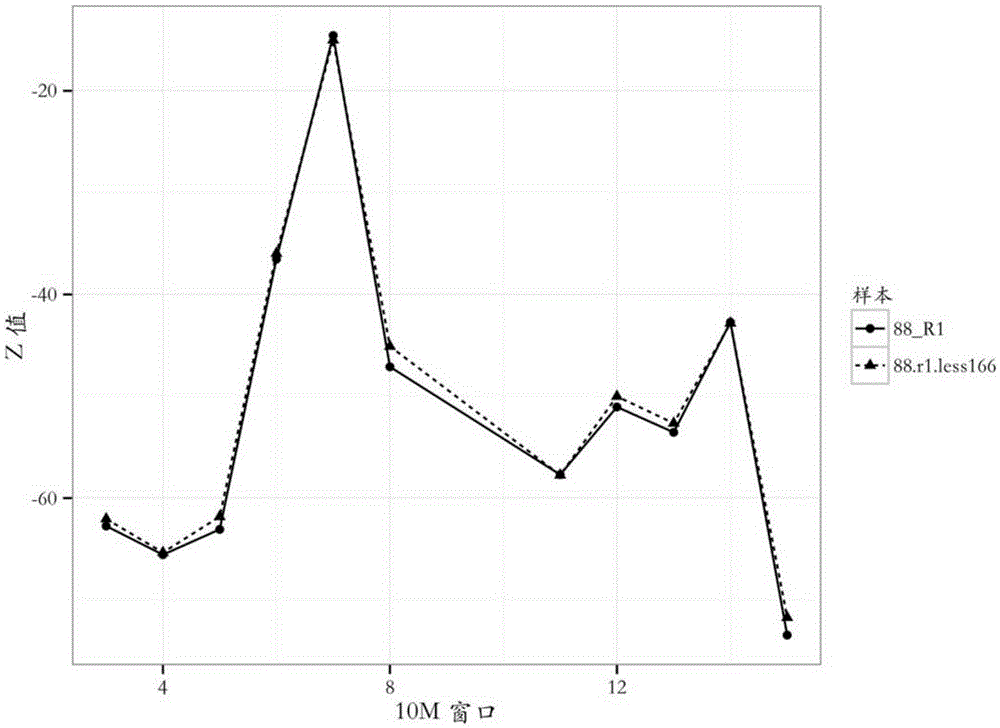
[0061] (7) Determine the source of cnv: use the sample M1 to simulate the Turner syndrome (XO) sample, such as figure 2 As shown, the deviation of the curve 88_R1 from the abscissa axis (z value = 0) indicates the presence of cnv. Combined with the curves 88_R1 and 88.r1.less166, it can be seen that the absolute value o...
Embodiment 3c
[0062] The judgment of embodiment 3cnv source fetus
[0063] (1) According to the method of step (1) of Example 1, the free plasma DNA of a pregnant woman pregnant with a fetus with trisomy 21 was extracted, and this group of samples was recorded as M2 as the sample to be tested.
[0064] (2) This step is the same as step (2) in Example 1.
[0065] (3) This step is the same as step (3) in Example 1.
[0066] (4) Align the DNA sequence of each sample to the human reference genome sequence (chromosome 21).
[0067] (5) This step is the same as step (5) in Example 2.
[0068] (6) This step is the same as step (6) in Example 2.
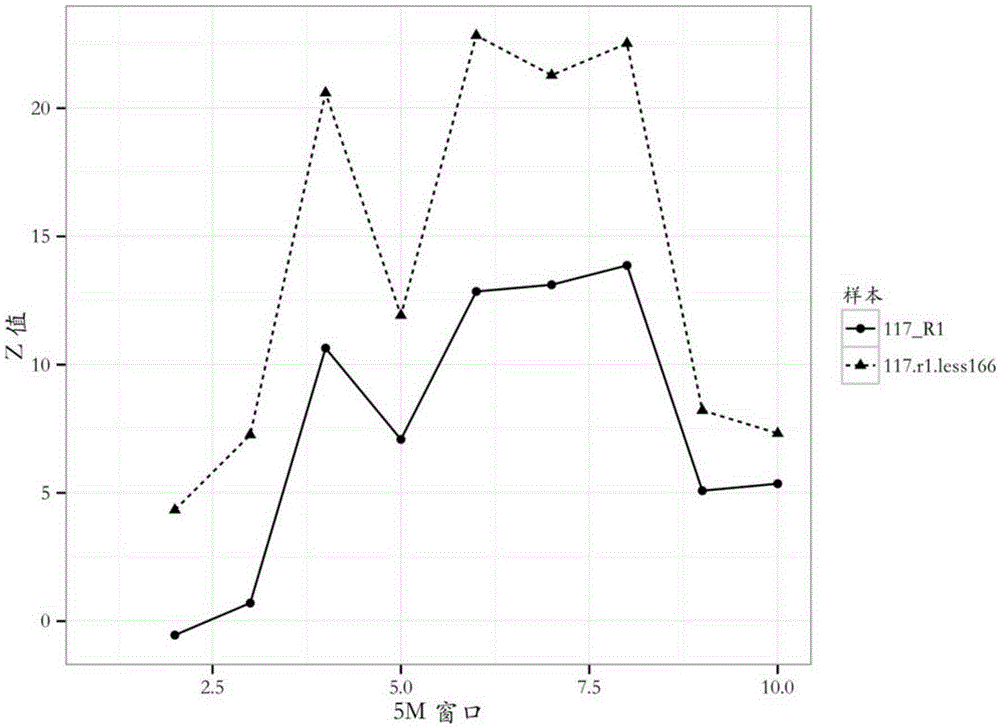
[0069] (7) cnv source determination: if image 3 As shown, the deviation of the curve 117_R1 from the abscissa axis (z value = 0) indicates the presence of cnv. Combined with the curves 117_R1 and 117.r1.less166, it can be seen that the absolute value of the z values of the two groups of samples has a relationship that abs(117.r1.less166) is greate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com