Omega-3 desaturase from Phytophthora nicotianae, vector containing desaturase, recombinant microorganism and their application
A technology for recombining microorganisms and Phytophthora parasitica, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Determination of an omega-3 desaturase with a normal temperature preference of 20C
[0026] Five known ω-3 desaturase sequences (genes OPIN17, sdd17, PsD17, PrD17 and PaD17) with a normal temperature preference of 20C were compared in the NCBI library, and two were screened out based on sequence similarity and homology analysis. The ω-3 desaturase gene sequences with high similarity and close relationship with the known sequences are from Phytophthora parasitica and Aphanomyces mediatedus respectively, and the corresponding gene numbers are XM_008906963 and XM_008870610 respectively. The annotations of these two sequences in the NCBI library belong to the family of fatty acid desaturases, but their specific functions have not been annotated in detail.
[0027] By ClustalW2 software, the above two sequences of the coding regions of the genes were compared with the aforementioned five known 20C-preferring ω-3 desaturase sequences, and it was found that the abov...
Embodiment 2
[0030]Embodiment 2: Construction of recombinant expression vector
[0031] 1. Obtain the target gene
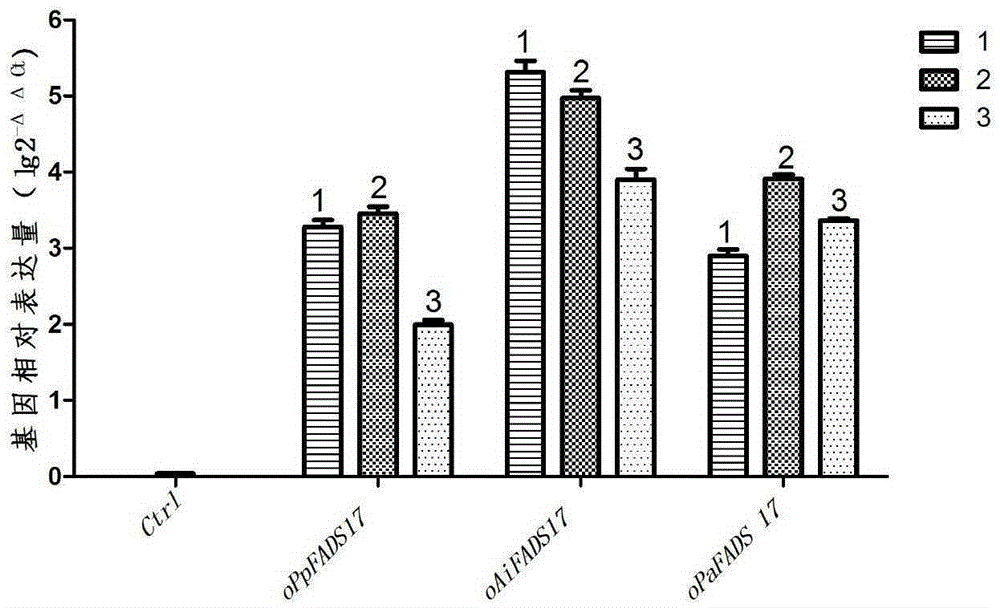
[0032] Since the codon usage bias of the gene sequences derived from Phytophthora parasitica, Aphanomyces meloni and Pythium melonica is different from that of the host Saccharomyces cerevisiae, according to the codon usage bias of eukaryotes, the nucleic acid sequences were analyzed using the GenscriptOptimumGeneTMsystem Codon optimization, artificially synthesized optimized gene sequences oPpFADS17, oAiFADS17 and oPaFADS17 (as positive controls), such as SEQIDNO. PUC57-oAiFADS17 and PUC57-oPaFADS17 were stored in Escherichia coli Top10 (purchased from GenScript, Nanjing, China).
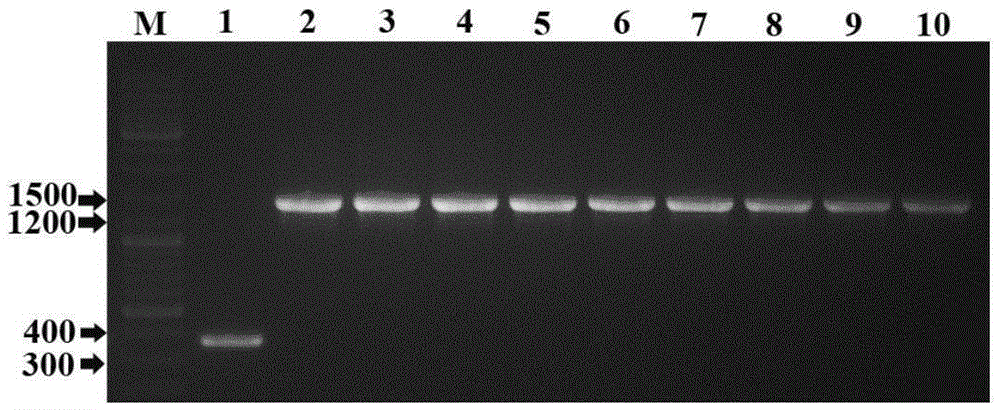
[0033] Primers were designed according to the sequences of oPpFADS17, oAiFADS17 and oPaFADS17, and restriction sites EcoRI and XhoI (underlined) were added to each pair of primers. Using the plasmids PUC57-oPpFADS17, PUC57-oAiFADS17, and PUC57-oPaFADS17 respectively containing the above optimize...
experiment example 4
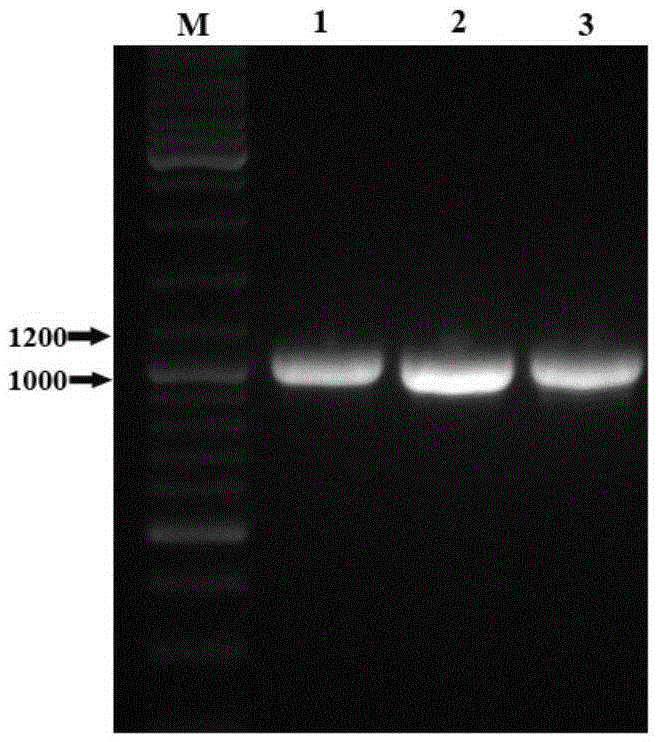
[0062] Experimental Example 4, PCR verification of Saccharomyces cerevisiae transformants
[0063] Pick the well-growth colony from the plate and inoculate it into 5mL YPD medium containing ampicillin resistance, culture at 200rpm at 30°C for 48h, extract the plasmid with a plasmid extraction kit (purchased from Tiangen Company), and measure A260nm and A280nm value, calculate the concentration of the plasmid, and store at low temperature.
[0064] The corresponding primers were used for PCR identification. Primers are as follows:
[0065] T7TAATACGACTCACTATAGGG
[0066] T7terminatorTCGGTTAGAGCGGATGTG
[0067] The PCR reaction system is as follows: ddH 2 O7 μL, 10×TaqMIX 10 μL, universal primer T71 μL, universal primer T7terminator 1 μL, template (plasmid) 1 μL. PCR reaction conditions: 94°C for 5min, 94°C for 30s, 58°C for 30s, 72°C for 1.5min, 30 cycles, 72°C for 7min. After the amplification reaction, take 3 μL of the PCR product and perform 1wt% agarose gel electropho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com