SNP molecular marker for identifying Shanghai colorful waxy corn NO.1 and identifying method thereof
A technology of molecular markers and waxy corn, which is applied in the field of biological identification, can solve the problems that the research has not yet been carried out, and achieve the effect of good repeatability, simple method, and less sample consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Determining SNP Molecular Markers
[0060] The SNP site sequence was obtained by 2b-RAD degenerate genome sequencing of Huwucaihuanuo 1, including 9 SNP site sequences SNP1-SNP9 of Huwucaihuanuo 1, and each SNP molecular marker sequence contained 1 Hu The SNP site of Wucaihuanuo 1. The nucleotide sequences of the molecular markers SNP1-SNP9 are respectively shown in SEQ ID NO.1-9, and the specific SNP molecular marker information is shown in Table 3.
Embodiment 2
[0061] Example 2 Variety identification of corn Huwucaihuanuo 1
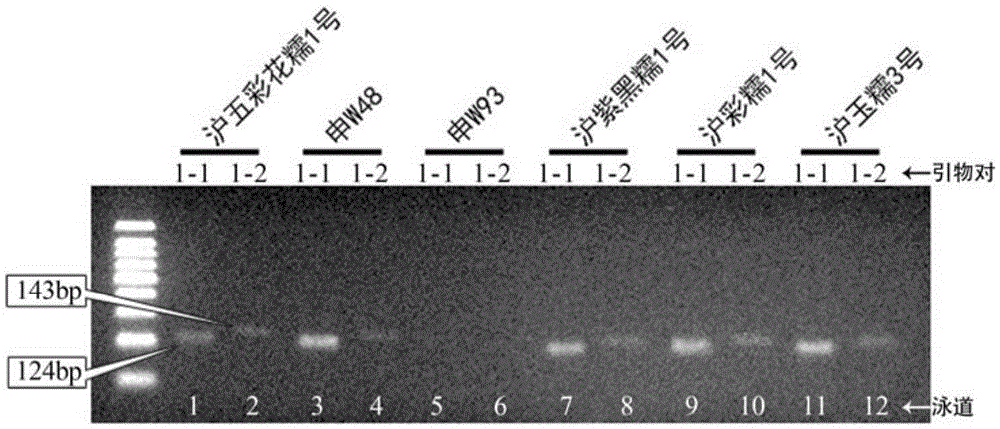
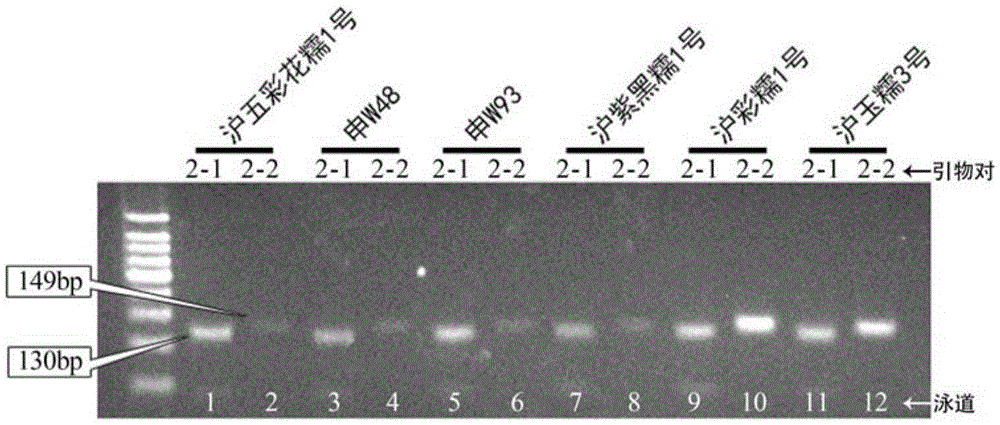
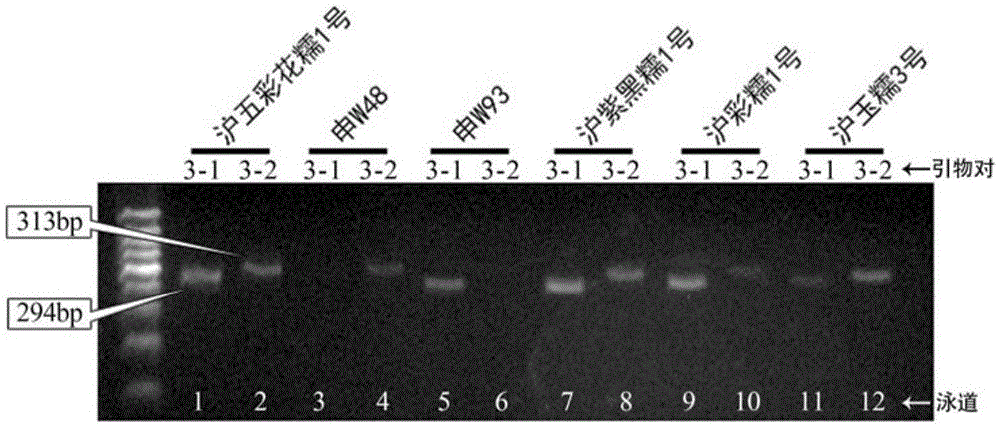
[0062] Using the combination of primers designed in this invention to carry out variety identification on genomic DNA samples of glutinous rice varieties Huwucaihuanuo 1, Shen W48, Shen W93, Huziheinuo 1, Hucainuo 1, and Huyunuo 3 and identification of germplasm resources.
[0063] (1) Design primers
[0064] According to the SNP molecular markers shown in Table 3 determined in Example 1, a pair of AC-PCR primers is designed, and the AC-PCR primers include primer pair combinations 1-9, and each primer pair combination includes two primer pairs, specifically See Table 2 for primer sequences;
[0065] (2) Genomic DNA extraction
[0066] The CTAB method is used to extract the genomic DNA of any organ or tissue of the samples to be identified: sample 1, sample 2, sample 3, sample 4, sample 5, sample 6.
[0067] (3) PCR amplification
[0068] Perform PCR amplification on the genomic DNA of the sample to be ident...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com